????????????
ベイズ推論による機械学習入門 須山 敦志 著 を読む
????????????
Variational inference for Bayesian DimensionalityReduction
????????????
コードの理解は後回しにして、どんどにこう。DimensionalityReductionモジュールを導入して、demoを実行へ:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 |
""" Variational inference for Bayesian DimensionalityReduction """ module DimensionalityReduction using Distributions #using ProgressMeter export DRModel export sample_data, VI #################### ## Types struct DRModel D::Int M::Int sigma2_y::Float64 m_W::Array{Float64, 2} # MxD Sigma_W::Array{Float64, 3} # MxMxD m_mu::Array{Float64, 1} # D Sigma_mu::Array{Float64, 2} # DxD end #################### ## functions function sqsum(mat::Array{Float64}, idx::Int) return squeeze(sum(mat, idx), idx) end """ Sample data given hyperparameters. """ function sample_data(N::Int, model::DRModel) D = model.D M = model.M W = zeros(M, D) mu = zeros(D) for d in 1 : D W[:,d] = rand(MvNormal(model.m_W[:,d], model.Sigma_W[:,:,d])) end mu = rand(MvNormal(model.m_mu, model.Sigma_mu)) Y = zeros(D, N) X = randn(M, N) for n in 1 : N Y[:,n] = rand(MvNormal(W'*X[:,n] + mu, model.sigma2_y*eye(D))) end return Y, X, W, mu end function init(Y::Array{Float64, 2}, prior::DRModel) M = prior.M D, N = size(Y) X = randn(M, N) XX = zeros(M, M, N) for n in 1 : N XX[:,:,n] = X[:,n]*X[:,n]' + eye(M) end return X, XX end function update_W(Y::Array{Float64, 2}, prior::DRModel, posterior::DRModel, X::Array{Float64, 2}, XX::Array{Float64, 3}) D = prior.D M = prior.M N = size(Y, 2) m_W = zeros(M, D) Sigma_W = zeros(M, M, D) mu = posterior.m_mu for d in 1 : D Sigma_W[:,:,d] = inv(inv(prior.sigma2_y)*sqsum(XX, 3) + inv(prior.Sigma_W[:,:,d])) m_W[:,d] = Sigma_W[:,:,d]*(inv(prior.sigma2_y)*X*(Y[[d],:] - mu[d]*ones(1, N))' + inv(prior.Sigma_W[:,:,d])*prior.m_W[:,d]) end return DRModel(D, M, prior.sigma2_y, m_W, Sigma_W, posterior.m_mu, posterior.Sigma_mu) end function update_mu(Y::Array{Float64, 2}, prior::DRModel, posterior::DRModel, X::Array{Float64, 2}, XX::Array{Float64, 3}) N = size(Y, 2) D = prior.D M = prior.M W = posterior.m_W Sigma_mu = inv(N*inv(prior.sigma2_y)*eye(D) + inv(prior.Sigma_mu)) m_mu = Sigma_mu*(inv(prior.sigma2_y)*sqsum(Y - W'*X, 2) + inv(prior.Sigma_mu)*prior.m_mu) return DRModel(D, M, prior.sigma2_y, posterior.m_W, posterior.Sigma_W, m_mu, Sigma_mu) end function update_X(Y::Array{Float64, 2}, posterior::DRModel) D, N = size(Y) M = posterior.M W = posterior.m_W WW = zeros(M, M, D) for d in 1 : D WW[:,:,d] = W[:,d]*W[:,d]' + posterior.Sigma_W[:,:,d] end mu = posterior.m_mu X = zeros(M, N) XX = zeros(M, M, N) for n in 1 : N Sigma = inv(inv(posterior.sigma2_y)*sqsum(WW, 3) + eye(M)) X[:,n] = inv(posterior.sigma2_y)*Sigma*W*(Y[:,n] - mu) XX[:,:,n] = X[:,n] * X[:,n]' + Sigma end return X, XX end function interpolate(mask::BitArray{2}, X::Array{Float64, 2}, posterior::DRModel) Y_est = posterior.m_W'*X + repmat(posterior.m_mu, 1, size(X, 2)) return return Y_est[mask] end """ Compute variational posterior distributions. """ function VI(Y::Array{Float64, 2}, prior::DRModel, max_iter::Int) X, XX = init(Y, prior) mask = isnan.(Y) sum_nan = sum(mask) posterior = deepcopy(prior) #progress = Progress(max_iter) for iter in 1 : max_iter # progress #next!(progress) # Interpolate if sum_nan > 0 Y[mask] = interpolate(mask, X, posterior) end # M-step posterior = update_W(Y, prior, posterior, X, XX) posterior = update_mu(Y, prior, posterior, X, XX) # E-step X, XX = update_X(Y, posterior) end return posterior, X end end |
Out: DimensionalityReduction
|
1 |
using DimensionalityReduction |
では、以下、demo_DimensionalityReduction.jlの実行へ:
ただしsklearnをpyimportするので、事前にアナコンダからscikit-learnをクリックしてインストールしておく必要あり。
|
1 2 3 4 5 |
################################### ## Demo script for Bayesian Dimensionality Reduction using PyPlot, PyCall @pyimport sklearn.datasets as datasets |
|
1 2 |
push!(LOAD_PATH,"/Users/******/Desktop/BayesBook-master/src/DimensionalityReduction/src") import DimensionalityReduction |
|
1 2 3 4 5 6 7 8 9 10 11 12 |
function load_facedata(skip::Int) face = datasets.fetch_olivetti_faces() Y_raw = face["images"] N, S_raw, _ = size(Y_raw) L = round(Int, S_raw / skip) Y_tmp = Y_raw[:,1:skip:end, 1:skip:end] Y = convert(Array{Float64, 2}, reshape(Y_tmp, N, size(Y_tmp,2)*size(Y_tmp,3))') D = size(Y, 1) return Y, D, L end |
Out: load_facedata (generic function with 1 method)
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
function visualize(Y::Array{Float64,2}, L::Int) D, N = size(Y) base = round(Int, sqrt(N)) v = round(Int, (L*ceil(N / base))) h = L * base pic = zeros(v, h) for n in 1 : N i = round(Int, (L*ceil(n / base))) idx1 = i - L + 1 : i idx2 = L*mod(n-1, base)+1 : L*(mod(n-1, base) + 1) pic[idx1,idx2] = reshape(Y[:,n], L, L) end imshow(pic, cmap=ColorMap("gray")) end |
Out: visualize (generic function with 1 method)
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 |
function visualize(Y::Array{Float64,2}, L::Int, mask::BitArray{2}) # for missing D, N = size(Y) base = round(Int, sqrt(N)) v = round(Int, (L*ceil(N / base))) h = L * base pic = zeros(v, h, 3) Y_3dim = zeros(D, N, 3) for i in 1 : 3 if i == 2 Y_tmp = deepcopy(Y) Y_tmp[mask] = 1 Y_3dim[:,:,i] = Y_tmp else Y_tmp = deepcopy(Y) Y_tmp[mask] = 0 Y_3dim[:,:,i] = Y_tmp end end for n in 1 : N i = round(Int, (L*ceil(n / base))) idx1 = i - L + 1 : i idx2 = L*mod(n-1, base)+1 : L*(mod(n-1, base) + 1) for i in 1 : 3 pic[idx1,idx2,i] = reshape(Y_3dim[:,n,i], L, L) end end imshow(pic, cmap=ColorMap("gray")) end |
Out: visualize (generic function with 2 methods)
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 |
""" Run a demo script of missing data interpolation for face dataset. """ function test_face_missing() # load data skip = 2 Y, D, L = load_facedata(skip) # mask missing_rate = 0.50 mask = rand(size(Y)) .< missing_rate Y_obs = deepcopy(Y) Y_obs[mask] = NaN # known parames M = 16 sigma2_y = 0.001 Sigma_W = zeros(M,M,D) Sigma_mu = 1.0 * eye(D) for d in 1 : D Sigma_W[:,:,d] = 0.1 * eye(M) end prior = DimensionalityReduction.DRModel(D, M, sigma2_y, zeros(M, D), Sigma_W, zeros(D), Sigma_mu) # learn & generate max_iter = 100 posterior, X_est = DimensionalityReduction.VI(deepcopy(Y_obs), prior, max_iter) Y_est = posterior.m_W'*X_est + repmat(posterior.m_mu, 1, size(X_est, 2)) Y_itp = deepcopy(Y_obs) Y_itp[mask] = Y_est[mask] #visualize N_show = 4^2 figure("Observation") clf() visualize(Y_obs[:,1:N_show], L, mask[:,1:N_show]) title("Observation") #figure("Estimation") #clf() #visualize(Y_est[:,1:N_show], L) #title("Estimation") figure("Interpolation") clf() visualize(Y_itp[:,1:N_show], L) title("Interpolation") figure("Truth") clf() visualize(Y[:,1:N_show], L) title("Truth") show() end |
Out: test_face_missing
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 |
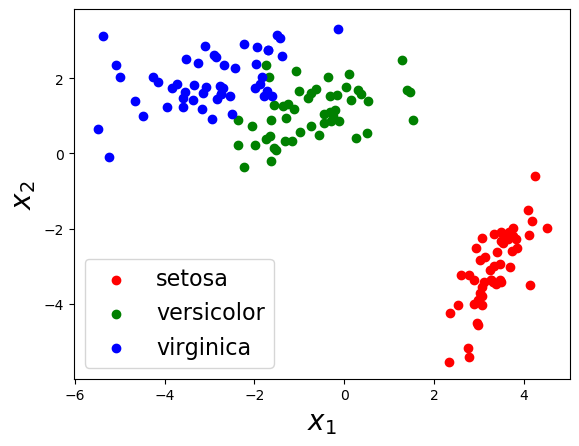
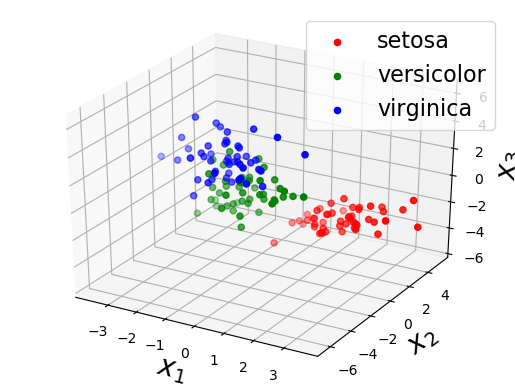
""" Run a dimensionality reduction demo using Iris dataset. """ function test_iris() ################## # load data iris = datasets.load_iris() Y_obs = iris["data"]' label_list = [iris["target_names"][elem+1] for elem in iris["target"]] D, N = size(Y_obs) ################## # 2D compression # model M = 2 sigma2_y = 0.001 Sigma_W = zeros(M,M,D) Sigma_mu = 1.0 * eye(D) for d in 1 : D Sigma_W[:,:,d] = 0.1 * eye(M) end prior = DimensionalityReduction.DRModel(D, M, sigma2_y, zeros(M, D), Sigma_W, zeros(D), Sigma_mu) # learn & generate max_iter = 100 posterior, X_est = DimensionalityReduction.VI(deepcopy(Y_obs), prior, max_iter) # visualize figure("2D plot") clf() scatter(X_est[1,1:50], X_est[2,1:50], color="r") scatter(X_est[1,51:100], X_est[2,51:100], color="g") scatter(X_est[1,101:end], X_est[2,101:end], color="b") xlabel("\$x_1\$", fontsize=20) ylabel("\$x_2\$", fontsize=20) legend([label_list[1], label_list[51], label_list[101]], fontsize=16) ################## # 3D compression # model M = 3 sigma2_y = 0.001 Sigma_W = zeros(M,M,D) Sigma_mu = 1.0 * eye(D) for d in 1 : D Sigma_W[:,:,d] = 0.1 * eye(M) end prior = DimensionalityReduction.DRModel(D, M, sigma2_y, zeros(M, D), Sigma_W, zeros(D), Sigma_mu) # learn & generate max_iter = 100 posterior, X_est = DimensionalityReduction.VI(deepcopy(Y_obs), prior, max_iter) # visualize figure("3D plot") clf() scatter3D(X_est[1,1:50], X_est[2,1:50], X_est[3,1:50], c="r") scatter3D(X_est[1,51:100], X_est[2,51:100], X_est[3,51:100], c="g") scatter3D(X_est[1,101:end], X_est[2,101:end], X_est[3,101:end], c="b") legend([label_list[1], label_list[51], label_list[101]], fontsize=16) xlabel("\$x_1\$", fontsize=20) ylabel("\$x_2\$", fontsize=20) zlabel("\$x_3\$", fontsize=20) show() end |
Out: test_iris
|
1 2 |
#test_face_missing() test_iris() |