COVID-19重症呼吸不全患者に対するHNFC/MVの治療選択に対する影響因子:因果分析=>ベイジアンネットワーク 16データでの解析
ROXの閾値を4.0から7.0まで0.1刻みで変動させてみて、人工呼吸率や死亡率予測への影響を調べる。
まずはデータファイルをデータフレームに読み込む。
|
1 2 3 4 |
#install.packages("tidyverse") library("tidyverse") df <- read.csv("data_f16R.csv", header=TRUE, fileEncoding="utf-8") |
次に31個のデータフレームにコピーして、おく。
またROX(4-7, 0.1刻み)とLIV(35.5に固定)の閾値の配列を作成する。
|
1 2 3 4 5 6 7 8 9 10 11 |
for(x in 1:31){assign(sprintf("df%d", x), df)} ROX_TH <- lapply(1:31, function(x){3.9 + 0.1 * x}) LIV_TH <- lapply(1:31, function(x){35.5 + 0.0 * x}) ROX <- c(ROX_TH[1], ROX_TH[2], ROX_TH[3], ROX_TH[4], ROX_TH[5], ROX_TH[6], ROX_TH[7], ROX_TH[8], ROX_TH[9], ROX_TH[10], ROX_TH[11], ROX_TH[12], ROX_TH[13], ROX_TH[14], ROX_TH[15], ROX_TH[16], ROX_TH[17], ROX_TH[18], ROX_TH[19], ROX_TH[20], ROX_TH[21], ROX_TH[22], ROX_TH[23], ROX_TH[24], ROX_TH[25], ROX_TH[26], ROX_TH[27], ROX_TH[28], ROX_TH[29], ROX_TH[30], ROX_TH[31]) ROX |
4
4.1
4.2
4.3
4.4
……….
6.5
6.6
6.7
6.8
6.9
7
ROXとLIVに関して、閾値以下を”lo”, 閾値より高い値を”hi”に置き換える
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 |
df1$ROX[df1$ROX <= ROX_TH[1]] <- "lo" df1$ROX[df1$ROX != "lo"] <- "hi" df1$LIV[df1$LIV <= LIV_TH[1]] <- "lo" df1$LIV[df1$LIV != "lo"] <- "hi" df2$ROX[df2$ROX <= ROX_TH[2]] <- "lo" df2$ROX[df2$ROX != "lo"] <- "hi" df2$LIV[df2$LIV <= LIV_TH[2]] <- "lo" df2$LIV[df2$LIV != "lo"] <- "hi" df3$ROX[df3$ROX <= ROX_TH[3]] <- "lo" df3$ROX[df3$ROX != "lo"] <- "hi" df3$LIV[df3$LIV <= LIV_TH[3]] <- "lo" df3$LIV[df3$LIV != "lo"] <- "hi" df4$ROX[df4$ROX <= ROX_TH[4]] <- "lo" df4$ROX[df4$ROX != "lo"] <- "hi" df4$LIV[df4$LIV <= LIV_TH[4]] <- "lo" df4$LIV[df4$LIV != "lo"] <- "hi" df5$ROX[df5$ROX <= ROX_TH[5]] <- "lo" df5$ROX[df5$ROX != "lo"] <- "hi" df5$LIV[df5$LIV <= LIV_TH[5]] <- "lo" df5$LIV[df5$LIV != "lo"] <- "hi" df6$ROX[df6$ROX <= ROX_TH[6]] <- "lo" df6$ROX[df6$ROX != "lo"] <- "hi" df6$LIV[df6$LIV <= LIV_TH[6]] <- "lo" df6$LIV[df6$LIV != "lo"] <- "hi" df7$ROX[df7$ROX <= ROX_TH[7]] <- "lo" df7$ROX[df7$ROX != "lo"] <- "hi" df7$LIV[df7$LIV <= LIV_TH[7]] <- "lo" df7$LIV[df7$LIV != "lo"] <- "hi" df8$ROX[df8$ROX <= ROX_TH[8]] <- "lo" df8$ROX[df8$ROX != "lo"] <- "hi" df8$LIV[df8$LIV <= LIV_TH[8]] <- "lo" df8$LIV[df8$LIV != "lo"] <- "hi" df9$ROX[df9$ROX <= ROX_TH[9]] <- "lo" df9$ROX[df9$ROX != "lo"] <- "hi" df9$LIV[df9$LIV <= LIV_TH[9]] <- "lo" df9$LIV[df9$LIV != "lo"] <- "hi" df10$ROX[df10$ROX <= ROX_TH[10]] <- "lo" df10$ROX[df10$ROX != "lo"] <- "hi" df10$LIV[df10$LIV <= LIV_TH[10]] <- "lo" df10$LIV[df10$LIV != "lo"] <- "hi" df11$ROX[df11$ROX <= ROX_TH[11]] <- "lo" df11$ROX[df11$ROX != "lo"] <- "hi" df11$LIV[df11$LIV <= LIV_TH[11]] <- "lo" df11$LIV[df11$LIV != "lo"] <- "hi" df12$ROX[df12$ROX <= ROX_TH[12]] <- "lo" df12$ROX[df12$ROX != "lo"] <- "hi" df12$LIV[df12$LIV <= LIV_TH[12]] <- "lo" df12$LIV[df12$LIV != "lo"] <- "hi" df13$ROX[df13$ROX <= ROX_TH[13]] <- "lo" df13$ROX[df13$ROX != "lo"] <- "hi" df13$LIV[df13$LIV <= LIV_TH[13]] <- "lo" df13$LIV[df13$LIV != "lo"] <- "hi" df14$ROX[df14$ROX <= ROX_TH[14]] <- "lo" df14$ROX[df14$ROX != "lo"] <- "hi" df14$LIV[df14$LIV <= LIV_TH[14]] <- "lo" df14$LIV[df14$LIV != "lo"] <- "hi" df15$ROX[df15$ROX <= ROX_TH[15]] <- "lo" df15$ROX[df15$ROX != "lo"] <- "hi" df15$LIV[df15$LIV <= LIV_TH[15]] <- "lo" df15$LIV[df15$LIV != "lo"] <- "hi" df16$ROX[df16$ROX <= ROX_TH[16]] <- "lo" df16$ROX[df16$ROX != "lo"] <- "hi" df16$LIV[df16$LIV <= LIV_TH[16]] <- "lo" df16$LIV[df16$LIV != "lo"] <- "hi" df17$ROX[df17$ROX <= ROX_TH[17]] <- "lo" df17$ROX[df17$ROX != "lo"] <- "hi" df17$LIV[df17$LIV <= LIV_TH[17]] <- "lo" df17$LIV[df17$LIV != "lo"] <- "hi" df18$ROX[df18$ROX <= ROX_TH[18]] <- "lo" df18$ROX[df18$ROX != "lo"] <- "hi" df18$LIV[df18$LIV <= LIV_TH[18]] <- "lo" df18$LIV[df18$LIV != "lo"] <- "hi" df19$ROX[df19$ROX <= ROX_TH[19]] <- "lo" df19$ROX[df19$ROX != "lo"] <- "hi" df19$LIV[df19$LIV <= LIV_TH[19]] <- "lo" df19$LIV[df19$LIV != "lo"] <- "hi" df20$ROX[df20$ROX <= ROX_TH[20]] <- "lo" df20$ROX[df20$ROX != "lo"] <- "hi" df20$LIV[df20$LIV <= LIV_TH[20]] <- "lo" df20$LIV[df20$LIV != "lo"] <- "hi" df21$ROX[df21$ROX <= ROX_TH[21]] <- "lo" df21$ROX[df21$ROX != "lo"] <- "hi" df21$LIV[df21$LIV <= LIV_TH[21]] <- "lo" df21$LIV[df21$LIV != "lo"] <- "hi" df22$ROX[df22$ROX <= ROX_TH[22]] <- "lo" df22$ROX[df22$ROX != "lo"] <- "hi" df22$LIV[df22$LIV <= LIV_TH[22]] <- "lo" df22$LIV[df22$LIV != "lo"] <- "hi" df23$ROX[df23$ROX <= ROX_TH[23]] <- "lo" df23$ROX[df23$ROX != "lo"] <- "hi" df23$LIV[df23$LIV <= LIV_TH[23]] <- "lo" df23$LIV[df23$LIV != "lo"] <- "hi" df24$ROX[df24$ROX <= ROX_TH[24]] <- "lo" df24$ROX[df24$ROX != "lo"] <- "hi" df24$LIV[df24$LIV <= LIV_TH[24]] <- "lo" df24$LIV[df24$LIV != "lo"] <- "hi" df25$ROX[df25$ROX <= ROX_TH[25]] <- "lo" df25$ROX[df25$ROX != "lo"] <- "hi" df25$LIV[df25$LIV <= LIV_TH[25]] <- "lo" df25$LIV[df25$LIV != "lo"] <- "hi" df26$ROX[df26$ROX <= ROX_TH[26]] <- "lo" df26$ROX[df26$ROX != "lo"] <- "hi" df26$LIV[df26$LIV <= LIV_TH[26]] <- "lo" df26$LIV[df26$LIV != "lo"] <- "hi" df27$ROX[df27$ROX <= ROX_TH[27]] <- "lo" df27$ROX[df27$ROX != "lo"] <- "hi" df27$LIV[df27$LIV <= LIV_TH[27]] <- "lo" df27$LIV[df27$LIV != "lo"] <- "hi" df28$ROX[df28$ROX <= ROX_TH[28]] <- "lo" df28$ROX[df28$ROX != "lo"] <- "hi" df28$LIV[df28$LIV <= LIV_TH[28]] <- "lo" df28$LIV[df28$LIV != "lo"] <- "hi" df29$ROX[df29$ROX <= ROX_TH[29]] <- "lo" df29$ROX[df29$ROX != "lo"] <- "hi" df29$LIV[df29$LIV <= LIV_TH[29]] <- "lo" df29$LIV[df29$LIV != "lo"] <- "hi" df30$ROX[df30$ROX <= ROX_TH[30]] <- "lo" df30$ROX[df30$ROX != "lo"] <- "hi" df30$LIV[df30$LIV <= LIV_TH[30]] <- "lo" df30$LIV[df30$LIV != "lo"] <- "hi" df31$ROX[df31$ROX <= ROX_TH[31]] <- "lo" df31$ROX[df31$ROX != "lo"] <- "hi" df31$LIV[df31$LIV <= LIV_TH[31]] <- "lo" df31$LIV[df31$LIV != "lo"] <- "hi" |
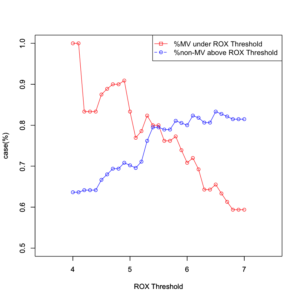
ROX閾値以下で人工呼吸有り(MV=ON)となった件数と、ROX閾値以下の件数から、ROX閾値以下での人工呼吸率を算定。
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 |
m1 = as.numeric(count(subset(df1, df1$MV == "on" & df1$ROX == "lo"))) n1 = as.numeric(count(subset(df1, df1$ROX == "lo"))) r1 <- as.numeric(m1/n1) m2 = as.numeric(count(subset(df2, df2$MV == "on" & df2$ROX == "lo"))) n2 = as.numeric(count(subset(df2, df2$ROX == "lo"))) r2 <- as.numeric(m2/n2) m3 = as.numeric(count(subset(df3, df3$MV == "on" & df3$ROX == "lo"))) n3 = as.numeric(count(subset(df3, df3$ROX == "lo"))) r3 <- as.numeric(m3/n3) m4 = as.numeric(count(subset(df4, df4$MV == "on" & df4$ROX == "lo"))) n4 = as.numeric(count(subset(df4, df4$ROX == "lo"))) r4 <- as.numeric(m4/n4) m5 = as.numeric(count(subset(df5, df5$MV == "on" & df5$ROX == "lo"))) n5 = as.numeric(count(subset(df5, df5$ROX == "lo"))) r5 <- as.numeric(m5/n5) m6 = as.numeric(count(subset(df6, df6$MV == "on" & df6$ROX == "lo"))) n6 = as.numeric(count(subset(df6, df6$ROX == "lo"))) r6 <- as.numeric(m6/n6) m7 = as.numeric(count(subset(df7, df7$MV == "on" & df7$ROX == "lo"))) n7 = as.numeric(count(subset(df7, df7$ROX == "lo"))) r7 <- as.numeric(m7/n7) m8 = as.numeric(count(subset(df8, df8$MV == "on" & df8$ROX == "lo"))) n8 = as.numeric(count(subset(df8, df8$ROX == "lo"))) r8 <- as.numeric(m8/n8) m9 = as.numeric(count(subset(df9, df9$MV == "on" & df9$ROX == "lo"))) n9 = as.numeric(count(subset(df9, df9$ROX == "lo"))) r9 <- as.numeric(m9/n9) m10 = as.numeric(count(subset(df10, df10$MV == "on" & df10$ROX == "lo"))) n10 = as.numeric(count(subset(df10, df10$ROX == "lo"))) r10 <- as.numeric(m10/n10) m11 = as.numeric(count(subset(df11, df11$MV == "on" & df11$ROX == "lo"))) n11 = as.numeric(count(subset(df11, df11$ROX == "lo"))) r11 <- as.numeric(m11/n11) m12 = as.numeric(count(subset(df12, df12$MV == "on" & df12$ROX == "lo"))) n12 = as.numeric(count(subset(df12, df12$ROX == "lo"))) r12 <- as.numeric(m12/n12) m13 = as.numeric(count(subset(df13, df13$MV == "on" & df13$ROX == "lo"))) n13 = as.numeric(count(subset(df13, df13$ROX == "lo"))) r13 <- as.numeric(m13/n13) m14 = as.numeric(count(subset(df14, df14$MV == "on" & df14$ROX == "lo"))) n14 = as.numeric(count(subset(df14, df14$ROX == "lo"))) r14 <- as.numeric(m14/n14) m15 = as.numeric(count(subset(df15, df15$MV == "on" & df15$ROX == "lo"))) n15 = as.numeric(count(subset(df15, df15$ROX == "lo"))) r15 <- as.numeric(m15/n15) m16 = as.numeric(count(subset(df16, df16$MV == "on" & df16$ROX == "lo"))) n16 = as.numeric(count(subset(df16, df16$ROX == "lo"))) r16 <- as.numeric(m16/n16) m17 = as.numeric(count(subset(df17, df17$MV == "on" & df17$ROX == "lo"))) n17 = as.numeric(count(subset(df17, df17$ROX == "lo"))) r17 <- as.numeric(m17/n17) m18 = as.numeric(count(subset(df18, df18$MV == "on" & df18$ROX == "lo"))) n18 = as.numeric(count(subset(df18, df18$ROX == "lo"))) r18 <- as.numeric(m18/n18) m19 = as.numeric(count(subset(df19, df19$MV == "on" & df19$ROX == "lo"))) n19 = as.numeric(count(subset(df19, df19$ROX == "lo"))) r19 <- as.numeric(m19/n19) m20 = as.numeric(count(subset(df20, df20$MV == "on" & df20$ROX == "lo"))) n20 = as.numeric(count(subset(df20, df20$ROX == "lo"))) r20 <- as.numeric(m20/n20) m21 = as.numeric(count(subset(df21, df21$MV == "on" & df21$ROX == "lo"))) n21 = as.numeric(count(subset(df21, df21$ROX == "lo"))) r21 <- as.numeric(m21/n21) m22 = as.numeric(count(subset(df22, df22$MV == "on" & df22$ROX == "lo"))) n22 = as.numeric(count(subset(df22, df22$ROX == "lo"))) r22 <- as.numeric(m22/n22) m23 = as.numeric(count(subset(df23, df23$MV == "on" & df23$ROX == "lo"))) n23 = as.numeric(count(subset(df23, df23$ROX == "lo"))) r23 <- as.numeric(m23/n23) m24 = as.numeric(count(subset(df24, df24$MV == "on" & df24$ROX == "lo"))) n24 = as.numeric(count(subset(df24, df24$ROX == "lo"))) r24 <- as.numeric(m24/n24) m25 = as.numeric(count(subset(df25, df25$MV == "on" & df25$ROX == "lo"))) n25 = as.numeric(count(subset(df25, df25$ROX == "lo"))) r25 <- as.numeric(m25/n25) m26 = as.numeric(count(subset(df26, df26$MV == "on" & df26$ROX == "lo"))) n26 = as.numeric(count(subset(df26, df26$ROX == "lo"))) r26 <- as.numeric(m26/n26) m27 = as.numeric(count(subset(df27, df27$MV == "on" & df27$ROX == "lo"))) n27 = as.numeric(count(subset(df27, df27$ROX == "lo"))) r27 <- as.numeric(m27/n27) m28 = as.numeric(count(subset(df28, df28$MV == "on" & df28$ROX == "lo"))) n28 = as.numeric(count(subset(df28, df28$ROX == "lo"))) r28 <- as.numeric(m28/n28) m29 = as.numeric(count(subset(df29, df29$MV == "on" & df29$ROX == "lo"))) n29 = as.numeric(count(subset(df29, df29$ROX == "lo"))) r29 <- as.numeric(m29/n29) m30 = as.numeric(count(subset(df30, df30$MV == "on" & df30$ROX == "lo"))) n30 = as.numeric(count(subset(df30, df30$ROX == "lo"))) r30 <- as.numeric(m30/n30) m31 = as.numeric(count(subset(df31, df31$MV == "on" & df31$ROX == "lo"))) n31 = as.numeric(count(subset(df31, df31$ROX == "lo"))) r31 <- as.numeric(m31/n31) |
ROX閾値より高く人工呼吸無し(MV=OFF)となった件数と、ROX閾値より高い件数から、ROX閾値より高い非人工呼吸率を算定。
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 |
m1R = as.numeric(count(subset(df1, df1$MV == "off" & df1$ROX == "hi"))) n1R = as.numeric(count(subset(df1, df1$ROX == "hi"))) r1R <- as.numeric(m1R/n1R) m2R = as.numeric(count(subset(df2, df2$MV == "off" & df2$ROX == "hi"))) n2R = as.numeric(count(subset(df2, df2$ROX == "hi"))) r2R <- as.numeric(m2R/n2R) m3R = as.numeric(count(subset(df3, df3$MV == "off" & df3$ROX == "hi"))) n3R = as.numeric(count(subset(df3, df3$ROX == "hi"))) r3R <- as.numeric(m3R/n3R) m4R = as.numeric(count(subset(df4, df4$MV == "off" & df4$ROX == "hi"))) n4R = as.numeric(count(subset(df4, df4$ROX == "hi"))) r4R <- as.numeric(m4R/n4R) m5R = as.numeric(count(subset(df5, df5$MV == "off" & df5$ROX == "hi"))) n5R = as.numeric(count(subset(df5, df5$ROX == "hi"))) r5R <- as.numeric(m5R/n5R) m6R = as.numeric(count(subset(df6, df6$MV == "off" & df6$ROX == "hi"))) n6R = as.numeric(count(subset(df6, df6$ROX == "hi"))) r6R <- as.numeric(m6R/n6R) m7R = as.numeric(count(subset(df7, df7$MV == "off" & df7$ROX == "hi"))) n7R = as.numeric(count(subset(df7, df7$ROX == "hi"))) r7R <- as.numeric(m7R/n7R) m8R = as.numeric(count(subset(df8, df8$MV == "off" & df8$ROX == "hi"))) n8R = as.numeric(count(subset(df8, df8$ROX == "hi"))) r8R <- as.numeric(m8R/n8R) m9R = as.numeric(count(subset(df9, df9$MV == "off" & df9$ROX == "hi"))) n9R = as.numeric(count(subset(df9, df9$ROX == "hi"))) r9R <- as.numeric(m9R/n9R) m10R = as.numeric(count(subset(df10, df10$MV == "off" & df10$ROX == "hi"))) n10R = as.numeric(count(subset(df10, df10$ROX == "hi"))) r10R <- as.numeric(m10R/n10R) m11R = as.numeric(count(subset(df11, df11$MV == "off" & df11$ROX == "hi"))) n11R = as.numeric(count(subset(df11, df11$ROX == "hi"))) r11R <- as.numeric(m11R/n11R) m12R = as.numeric(count(subset(df12, df12$MV == "off" & df12$ROX == "hi"))) n12R = as.numeric(count(subset(df12, df12$ROX == "hi"))) r12R <- as.numeric(m12R/n12R) m13R = as.numeric(count(subset(df13, df13$MV == "off" & df13$ROX == "hi"))) n13R = as.numeric(count(subset(df13, df13$ROX == "hi"))) r13R <- as.numeric(m13R/n13R) m14R = as.numeric(count(subset(df14, df14$MV == "off" & df14$ROX == "hi"))) n14R = as.numeric(count(subset(df14, df14$ROX == "hi"))) r14R <- as.numeric(m14R/n14R) m15R = as.numeric(count(subset(df15, df15$MV == "off" & df15$ROX == "hi"))) n15R = as.numeric(count(subset(df15, df15$ROX == "hi"))) r15R <- as.numeric(m15R/n15R) m16R = as.numeric(count(subset(df16, df16$MV == "off" & df16$ROX == "hi"))) n16R = as.numeric(count(subset(df16, df16$ROX == "hi"))) r16R <- as.numeric(m16R/n16R) m17R = as.numeric(count(subset(df17, df17$MV == "off" & df17$ROX == "hi"))) n17R = as.numeric(count(subset(df17, df17$ROX == "hi"))) r17R <- as.numeric(m17R/n17R) m18R = as.numeric(count(subset(df18, df18$MV == "off" & df18$ROX == "hi"))) n18R = as.numeric(count(subset(df18, df18$ROX == "hi"))) r18R <- as.numeric(m18R/n18R) m19R = as.numeric(count(subset(df19, df19$MV == "off" & df19$ROX == "hi"))) n19R = as.numeric(count(subset(df19, df19$ROX == "hi"))) r19R <- as.numeric(m19R/n19R) m20R = as.numeric(count(subset(df20, df20$MV == "off" & df20$ROX == "hi"))) n20R = as.numeric(count(subset(df20, df20$ROX == "hi"))) r20R <- as.numeric(m20R/n20R) m21R = as.numeric(count(subset(df21, df21$MV == "off" & df21$ROX == "hi"))) n21R = as.numeric(count(subset(df21, df21$ROX == "hi"))) r21R <- as.numeric(m21R/n21R) m22R = as.numeric(count(subset(df22, df22$MV == "off" & df22$ROX == "hi"))) n22R = as.numeric(count(subset(df22, df22$ROX == "hi"))) r22R <- as.numeric(m22R/n22R) m23R = as.numeric(count(subset(df23, df23$MV == "off" & df23$ROX == "hi"))) n23R = as.numeric(count(subset(df23, df23$ROX == "hi"))) r23R <- as.numeric(m23R/n23R) m24R = as.numeric(count(subset(df24, df24$MV == "off" & df24$ROX == "hi"))) n24R = as.numeric(count(subset(df24, df24$ROX == "hi"))) r24R <- as.numeric(m24R/n24R) m25R = as.numeric(count(subset(df25, df25$MV == "off" & df25$ROX == "hi"))) n25R = as.numeric(count(subset(df25, df25$ROX == "hi"))) r25R <- as.numeric(m25R/n25R) m26R = as.numeric(count(subset(df26, df26$MV == "off" & df26$ROX == "hi"))) n26R = as.numeric(count(subset(df26, df26$ROX == "hi"))) r26R <- as.numeric(m26R/n26R) m27R = as.numeric(count(subset(df27, df27$MV == "off" & df27$ROX == "hi"))) n27R = as.numeric(count(subset(df27, df27$ROX == "hi"))) r27R <- as.numeric(m27R/n27R) m28R = as.numeric(count(subset(df28, df28$MV == "off" & df28$ROX == "hi"))) n28R = as.numeric(count(subset(df28, df28$ROX == "hi"))) r28R <- as.numeric(m28R/n28R) m29R = as.numeric(count(subset(df29, df29$MV == "off" & df29$ROX == "hi"))) n29R = as.numeric(count(subset(df29, df29$ROX == "hi"))) r29R <- as.numeric(m29R/n29R) m30R = as.numeric(count(subset(df30, df30$MV == "off" & df30$ROX == "hi"))) n30R = as.numeric(count(subset(df30, df30$ROX == "hi"))) r30R <- as.numeric(m30R/n30R) m31R = as.numeric(count(subset(df31, df31$MV == "off" & df31$ROX == "hi"))) n31R = as.numeric(count(subset(df31, df31$ROX == "hi"))) r31R <- as.numeric(m31R/n31R) |
ROX閾値以下で死亡(MMortality=yes)となった件数と、ROX閾値以下の件数から、ROX閾値以下での死亡率を算定。
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 |
s1 = as.numeric(count(subset(df1, df1$Mortality == "yes" & df1$ROX == "lo"))) t1 = as.numeric(count(subset(df1, df1$ROX == "lo"))) u1 <- as.numeric(s1/t1) s2 = as.numeric(count(subset(df2, df2$Mortality == "yes" & df2$ROX == "lo"))) t2 = as.numeric(count(subset(df2, df2$ROX == "lo"))) u2 <- as.numeric(s2/t2) s3 = as.numeric(count(subset(df3, df3$Mortality == "yes" & df3$ROX == "lo"))) t3 = as.numeric(count(subset(df3, df3$ROX == "lo"))) u3 <- as.numeric(s3/t3) s4 = as.numeric(count(subset(df4, df4$Mortality == "yes" & df4$ROX == "lo"))) t4 = as.numeric(count(subset(df4, df4$ROX == "lo"))) u4 <- as.numeric(s4/t4) s5 = as.numeric(count(subset(df5, df5$Mortality == "yes" & df5$ROX == "lo"))) t5 = as.numeric(count(subset(df5, df5$ROX == "lo"))) u5 <- as.numeric(s5/t5) s6 = as.numeric(count(subset(df6, df6$Mortality == "yes" & df6$ROX == "lo"))) t6 = as.numeric(count(subset(df6, df6$ROX == "lo"))) u6 <- as.numeric(s6/t6) s7 = as.numeric(count(subset(df7, df7$Mortality == "yes" & df7$ROX == "lo"))) t7 = as.numeric(count(subset(df7, df7$ROX == "lo"))) u7 <- as.numeric(s7/t7) s8 = as.numeric(count(subset(df8, df8$Mortality == "yes" & df8$ROX == "lo"))) t8 = as.numeric(count(subset(df8, df8$ROX == "lo"))) u8 <- as.numeric(s8/t8) s9 = as.numeric(count(subset(df9, df9$Mortality == "yes" & df9$ROX == "lo"))) t9 = as.numeric(count(subset(df9, df9$ROX == "lo"))) u9 <- as.numeric(s9/t9) s10 = as.numeric(count(subset(df10, df10$Mortality == "yes" & df10$ROX == "lo"))) t10 = as.numeric(count(subset(df10, df10$ROX == "lo"))) u10 <- as.numeric(s10/t10) s11 = as.numeric(count(subset(df11, df11$Mortality == "yes" & df11$ROX == "lo"))) t11 = as.numeric(count(subset(df11, df11$ROX == "lo"))) u11 <- as.numeric(s11/n11) s12 = as.numeric(count(subset(df12, df12$Mortality == "yes" & df12$ROX == "lo"))) t12 = as.numeric(count(subset(df12, df12$ROX == "lo"))) u12 <- as.numeric(s12/t12) s13 = as.numeric(count(subset(df13, df13$Mortality == "yes" & df13$ROX == "lo"))) t13 = as.numeric(count(subset(df13, df13$ROX == "lo"))) u13 <- as.numeric(s13/t13) s14 = as.numeric(count(subset(df14, df14$Mortality == "yes" & df14$ROX == "lo"))) t14 = as.numeric(count(subset(df14, df14$ROX == "lo"))) u14 <- as.numeric(s14/t14) s15 = as.numeric(count(subset(df15, df15$Mortality == "yes" & df15$ROX == "lo"))) t15 = as.numeric(count(subset(df15, df15$ROX == "lo"))) u15 <- as.numeric(s15/t15) s16 = as.numeric(count(subset(df16, df16$Mortality == "yes" & df16$ROX == "lo"))) t16 = as.numeric(count(subset(df16, df16$ROX == "lo"))) u16 <- as.numeric(s16/t16) s17 = as.numeric(count(subset(df17, df17$Mortality == "yes" & df17$ROX == "lo"))) t17 = as.numeric(count(subset(df17, df17$ROX == "lo"))) u17 <- as.numeric(s17/t17) s18 = as.numeric(count(subset(df18, df18$Mortality == "yes" & df18$ROX == "lo"))) t18 = as.numeric(count(subset(df18, df18$ROX == "lo"))) u18 <- as.numeric(s18/t18) s19 = as.numeric(count(subset(df19, df19$Mortality == "yes" & df19$ROX == "lo"))) t19 = as.numeric(count(subset(df19, df19$ROX == "lo"))) u19 <- as.numeric(s19/t19) s20 = as.numeric(count(subset(df20, df20$Mortality == "yes" & df20$ROX == "lo"))) t20 = as.numeric(count(subset(df20, df20$ROX == "lo"))) u20 <- as.numeric(s20/t20) s21 = as.numeric(count(subset(df21, df21$Mortality == "yes" & df21$ROX == "lo"))) t21 = as.numeric(count(subset(df21, df21$ROX == "lo"))) u21 <- as.numeric(s21/t21) s22 = as.numeric(count(subset(df22, df22$Mortality == "yes" & df22$ROX == "lo"))) t22 = as.numeric(count(subset(df22, df22$ROX == "lo"))) u22 <- as.numeric(s22/t22) s23 = as.numeric(count(subset(df23, df23$Mortality == "yes" & df23$ROX == "lo"))) t23 = as.numeric(count(subset(df23, df23$ROX == "lo"))) u23 <- as.numeric(s23/t23) s24 = as.numeric(count(subset(df24, df24$Mortality == "yes" & df24$ROX == "lo"))) t24 = as.numeric(count(subset(df24, df24$ROX == "lo"))) u24 <- as.numeric(s24/t24) s25 = as.numeric(count(subset(df25, df25$Mortality == "yes" & df25$ROX == "lo"))) t25 = as.numeric(count(subset(df25, df25$ROX == "lo"))) u25 <- as.numeric(s25/t25) s26 = as.numeric(count(subset(df26, df26$Mortality == "yes" & df26$ROX == "lo"))) t26 = as.numeric(count(subset(df26, df26$ROX == "lo"))) u26 <- as.numeric(s26/t26) s27 = as.numeric(count(subset(df27, df27$Mortality == "yes" & df27$ROX == "lo"))) t27 = as.numeric(count(subset(df27, df27$ROX == "lo"))) u27 <- as.numeric(s27/t27) s28 = as.numeric(count(subset(df28, df28$Mortality == "yes" & df28$ROX == "lo"))) t28 = as.numeric(count(subset(df28, df28$ROX == "lo"))) u28 <- as.numeric(s28/t28) s29 = as.numeric(count(subset(df29, df29$Mortality == "yes" & df29$ROX == "lo"))) t29 = as.numeric(count(subset(df29, df29$ROX == "lo"))) u29 <- as.numeric(s29/t29) s30 = as.numeric(count(subset(df30, df30$Mortality == "yes" & df30$ROX == "lo"))) t30 = as.numeric(count(subset(df30, df30$ROX == "lo"))) u30 <- as.numeric(s30/t30) s31 = as.numeric(count(subset(df31, df31$Mortality == "yes" & df31$ROX == "lo"))) t31 = as.numeric(count(subset(df31, df31$ROX == "lo"))) u31 <- as.numeric(s31/t31) |
ROX閾値より高く生存(MMortality=no)となった件数と、ROX閾値より高い件数から、ROX閾値より高くての生存率を算定。
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 |
s1R = as.numeric(count(subset(df1, df1$Mortality == "no" & df1$ROX == "hi"))) t1R = as.numeric(count(subset(df1, df1$ROX == "hi"))) u1R <- as.numeric(s1R/t1R) s2R = as.numeric(count(subset(df2, df2$Mortality == "no" & df2$ROX == "hi"))) t2R = as.numeric(count(subset(df2, df2$ROX == "hi"))) u2R <- as.numeric(s2R/t2R) s3R = as.numeric(count(subset(df3, df3$Mortality == "no" & df3$ROX == "hi"))) t3R = as.numeric(count(subset(df3, df3$ROX == "hi"))) u3R <- as.numeric(s3R/t3R) s4R = as.numeric(count(subset(df4, df4$Mortality == "no" & df4$ROX == "hi"))) t4R = as.numeric(count(subset(df4, df4$ROX == "hi"))) u4R <- as.numeric(s4R/t4R) s5R = as.numeric(count(subset(df5, df5$Mortality == "no" & df5$ROX == "hi"))) t5R = as.numeric(count(subset(df5, df5$ROX == "hi"))) u5R <- as.numeric(s5R/t5R) s6R = as.numeric(count(subset(df6, df6$Mortality == "no" & df6$ROX == "hi"))) t6R = as.numeric(count(subset(df6, df6$ROX == "hi"))) u6R <- as.numeric(s6R/t6R) s7R = as.numeric(count(subset(df7, df7$Mortality == "no" & df7$ROX == "hi"))) t7R = as.numeric(count(subset(df7, df7$ROX == "hi"))) u7R <- as.numeric(s7R/t7R) s8R = as.numeric(count(subset(df8, df8$Mortality == "no" & df8$ROX == "hi"))) t8R = as.numeric(count(subset(df8, df8$ROX == "hi"))) u8R <- as.numeric(s8R/t8R) s9R = as.numeric(count(subset(df9, df9$Mortality == "no" & df9$ROX == "hi"))) t9R = as.numeric(count(subset(df9, df9$ROX == "hi"))) u9R <- as.numeric(s9R/t9R) s10R = as.numeric(count(subset(df10, df10$Mortality == "no" & df10$ROX == "hi"))) t10R = as.numeric(count(subset(df10, df10$ROX == "hi"))) u10R <- as.numeric(s10R/t10R) s11R = as.numeric(count(subset(df11, df11$Mortality == "no" & df11$ROX == "hi"))) t11R = as.numeric(count(subset(df11, df11$ROX == "hi"))) u11R <- as.numeric(s11R/n11R) s12R = as.numeric(count(subset(df12, df12$Mortality == "no" & df12$ROX == "hi"))) t12R = as.numeric(count(subset(df12, df12$ROX == "hi"))) u12R <- as.numeric(s12R/t12R) s13R = as.numeric(count(subset(df13, df13$Mortality == "no" & df13$ROX == "hi"))) t13R = as.numeric(count(subset(df13, df13$ROX == "hi"))) u13R <- as.numeric(s13R/t13R) s14R = as.numeric(count(subset(df14, df14$Mortality == "no" & df14$ROX == "hi"))) t14R = as.numeric(count(subset(df14, df14$ROX == "hi"))) u14R <- as.numeric(s14R/t14R) s15R = as.numeric(count(subset(df15, df15$Mortality == "no" & df15$ROX == "hi"))) t15R = as.numeric(count(subset(df15, df15$ROX == "hi"))) u15R <- as.numeric(s15R/t15R) s16R = as.numeric(count(subset(df16, df16$Mortality == "no" & df16$ROX == "hi"))) t16R = as.numeric(count(subset(df16, df16$ROX == "hi"))) u16R <- as.numeric(s16R/t16R) s17R = as.numeric(count(subset(df17, df17$Mortality == "no" & df17$ROX == "hi"))) t17R = as.numeric(count(subset(df17, df17$ROX == "hi"))) u17R <- as.numeric(s17R/t17R) s18R = as.numeric(count(subset(df18, df18$Mortality == "no" & df18$ROX == "hi"))) t18R = as.numeric(count(subset(df18, df18$ROX == "hi"))) u18R <- as.numeric(s18R/t18R) s19R = as.numeric(count(subset(df19, df19$Mortality == "no" & df19$ROX == "hi"))) t19R = as.numeric(count(subset(df19, df19$ROX == "hi"))) u19R <- as.numeric(s19R/t19R) s20R = as.numeric(count(subset(df20, df20$Mortality == "no" & df20$ROX == "hi"))) t20R = as.numeric(count(subset(df20, df20$ROX == "hi"))) u20R <- as.numeric(s20R/t20R) s21R = as.numeric(count(subset(df21, df21$Mortality == "no" & df21$ROX == "hi"))) t21R = as.numeric(count(subset(df21, df21$ROX == "hi"))) u21R <- as.numeric(s21R/t21R) s22R = as.numeric(count(subset(df22, df22$Mortality == "no" & df22$ROX == "hi"))) t22R = as.numeric(count(subset(df22, df22$ROX == "hi"))) u22R <- as.numeric(s22R/t22R) s23R = as.numeric(count(subset(df23, df23$Mortality == "no" & df23$ROX == "hi"))) t23R = as.numeric(count(subset(df23, df23$ROX == "hi"))) u23R <- as.numeric(s23R/t23R) s24R = as.numeric(count(subset(df24, df24$Mortality == "no" & df24$ROX == "hi"))) t24R = as.numeric(count(subset(df24, df24$ROX == "hi"))) u24R <- as.numeric(s24R/t24R) s25R = as.numeric(count(subset(df25, df25$Mortality == "no" & df25$ROX == "hi"))) t25R = as.numeric(count(subset(df25, df25$ROX == "hi"))) u25R <- as.numeric(s25R/t25R) s26R = as.numeric(count(subset(df26, df26$Mortality == "no" & df26$ROX == "hi"))) t26R = as.numeric(count(subset(df26, df26$ROX == "hi"))) u26R <- as.numeric(s26R/t26R) s27R = as.numeric(count(subset(df27, df27$Mortality == "no" & df27$ROX == "hi"))) t27R = as.numeric(count(subset(df27, df27$ROX == "hi"))) u27R <- as.numeric(s27R/t27R) s28R = as.numeric(count(subset(df28, df28$Mortality == "no" & df28$ROX == "hi"))) t28R = as.numeric(count(subset(df28, df28$ROX == "hi"))) u28R <- as.numeric(s28R/t28R) s29R = as.numeric(count(subset(df29, df29$Mortality == "no" & df29$ROX == "hi"))) t29R = as.numeric(count(subset(df29, df29$ROX == "hi"))) u29R <- as.numeric(s29R/t29R) s30R = as.numeric(count(subset(df30, df30$Mortality == "no" & df30$ROX == "hi"))) t30R = as.numeric(count(subset(df30, df30$ROX == "hi"))) u30R <- as.numeric(s30R/t30R) s31R = as.numeric(count(subset(df31, df31$Mortality == "no" & df31$ROX == "hi"))) t31R = as.numeric(count(subset(df31, df31$ROX == "hi"))) u31R <- as.numeric(s31R/t31R) |
それぞれの件数、率を配列にまとめる。
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 |
m_all <- c(m1, m2, m3, m4, m5, m6, m7, m8, m9, m10, m11, m12, m13, m14, m15, m16, m17, m18, m19, m20, m21, m22, m23, m24, m25, m26, m27, m28, m29, m30, m31) n_all <- c(n1, n2, n3, n4, n5, n6, n7, n8, n9, n10, n11, n12, n13, n14, n15, n16, n17, n18, n19, n20, n21, n22, n23, n24, n25, n26, n27, n28, n29, n30, n31) r_all <- c(r1, r2, r3, r4, r5, r6, r7, r8, r9, r10, r11, r12, r13, r14, r15, r16, r17, r18, r19, r20, r21, r22, r23, r24, r25, r26, r27, r28, r29, r30, r31) s_all <- c(s1, s2, s3, s4, s5, s6, s7, s8, s9, s10, s11, s12, s13, s14, s15, s16, s17, s18, s19, s20, s21, s22, s23, s24, s25, s26, s27, s28, s29, s30, s31) t_all <- c(t1, t2, t3, t4, t5, t6, t7, t8, t9, t10, t11, t12, t13, t14, t15, t16, t17, t18, t19, t20, t21, t22, t23, t24, t25, t26, t27, t28, t29, t30, t31) u_all <- c(u1, u2, u3, u4, u5, u6, u7, u8, u9, u10, u11, u12, u13, u14, u15, u16, u17, u18, u19, u20, u21, u22, u23, u24, u25, u26, u27, u28, u29, u30, u31) |
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 |
mR_all <- c(m1R, m2R, m3R, m4R, m5R, m6R, m7R, m8R, m9R, m10R, m11R, m12R, m13R, m14R, m15R, m16R, m17R, m18R, m19R, m20R, m21R, m22R, m23R, m24R, m25R, m26R, m27R, m28R, m29R, m30R, m31R) nR_all <- c(n1R, n2R, n3R, n4R, n5R, n6R, n7R, n8R, n9R, n10R, n11R, n12R, n13R, n14R, n15R, n16R, n17R, n18R, n19R, n20R, n21R, n22R, n23R, n24R, n25R, n26R, n27R, n28R, n29R, n30R, n31R) rR_all <- c(r1R, r2R, r3R, r4R, r5R, r6R, r7R, r8R, r9R, r10R, r11R, r12R, r13R, r14R, r15R, r16R, r17R, r18R, r19R, r20R, r21R, r22R, r23R, r24R, r25R, r26R, r27R, r28R, r29R, r30R, r31R) sR_all <- c(s1R, s2R, s3R, s4R, s5R, s6R, s7R, s8R, s9R, s10R, s11R, s12R, s13R, s14R, s15R, s16R, s17R, s18R, s19R, s20R, s21R, s22R, s23R, s24R, s25R, s26R, s27R, s28R, s29R, s30R, s31R) tR_all <- c(t1R, t2R, t3R, t4R, t5R, t6R, t7R, t8R, t9R, t10R, t11R, t12R, t13R, t14R, t15R, t16R, t17R, t18R, t19R, t20R, t21R, t22R, t23R, t24R, t25R, t26R, t27R, t28R, t29R, t30R, t31R) uR_all <- c(u1R, u2R, u3R, u4R, u5R, u6R, u7R, u8R, u9R, u10R, u11R, u12R, u13R, u14R, u15R, u16R, u17R, u18R, u19R, u20R, u21R, u22R, u23R, u24R, u25R, u26R, u27R, u28R, u29R, u30R, u31R) |
上記の配列をデータフレームdataに集める
|
1 2 |
data <- cbind(ROX, m_all, n_all, r_all, s_all, t_all, u_all, mR_all, nR_all, rR_all, sR_all, tR_all, uR_all) names(data) <- c("ROX", "m", "n", "r", "s", "t", "u", "mR", "nR", "rR", "sR", "tR", "uR") |
完成したデータフレームを表示
|
1 |
data |
A matrix: 31 × 13
ROX m_all n_all r_all s_all t_all u_all mR_all nR_all rR_all sR_all tR_all uR_all
4 4 4 1 1 4 0.25 35 55 0.6363636 52 55 0.9454545
4.1 4 4 1 1 4 0.25 35 55 0.6363636 52 55 0.9454545
4.2 5 6 0.8333333 1 6 0.1666667 34 53 0.6415094 50 53 0.9433962
…………………..
|
1 2 3 4 |
plot(data[,1], data[,4], type = "o", xlim =c(3.5,7.5), ylim=c(0.5, 1), col="red", xlab="ROX Threshold", ylab="case(%)") par(new=T) plot(data[,1], data[,10], type = "o", xlim =c(3.5,7.5), ylim=c(0.5, 1), col="blue", xlab="ROX Threshold", ylab="case(%)") legend("topright", legend = c("%MV under ROX Threshold","%non-MV above ROX Threshold"), col = c("red", "blue"), pch = c(0, 1),lty = c("solid", "dashed")) |
|
1 2 3 4 |
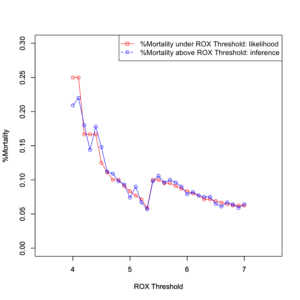
plot(data[,1], data[,7], type = "o", xlim =c(3.5,7.5), ylim=c(0.0, 1), col="red", xlab="ROX Threshold", ylab="case(%)") par(new=T) plot(data[,1], data[,13], type = "o", xlim =c(3.5,7.5), ylim=c(0.0, 1), col="blue", xlab="ROX Threshold", ylab="case(%)") legend("center", legend = c("%Mortality under ROX Threshold","%Survival above ROX Threshold"), col = c("red", "blue"), pch = c(0, 1),lty = c("solid", "dashed")) |
|
1 2 3 4 |
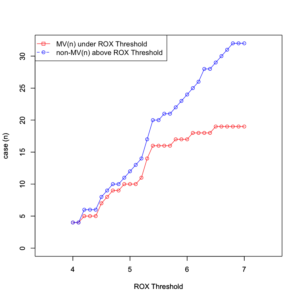
plot(data[,1], data[,2], type = "o", xlim =c(3.5,7.5), ylim=c(0, 32), col="red", xlab="ROX Threshold", ylab="case (n)") par(new=T) plot(data[,1], data[,3], type = "o", xlim =c(3.5,7.5), ylim=c(0, 32), col="blue", xlab="ROX Threshold", ylab="case (n)") legend("topleft", legend = c("MV(n) under ROX Threshold","non-MV(n) above ROX Threshold"), col = c("red", "blue"), pch = c(0, 1),lty = c("solid", "dashed")) |
|
1 2 3 4 |
plot(data[,1], data[,5], type = "o", xlim =c(3.5,7.5), ylim=c(0, 32), col="red", xlab="ROX Threshold", ylab="case (n)") par(new=T) plot(data[,1], data[,6], type = "o", xlim =c(3.5,7.5), ylim=c(0, 32), col="blue", xlab="ROX Threshold", ylab="case (n)") legend("topleft", legend = c("Mortality(n) under ROX Threshold","Survival(n) above ROX Threshold"), col = c("red", "blue"), pch = c(0, 1),lty = c("solid", "dashed")) |
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 |
df1F <- df1 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df1F <- df1F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df1F <- df1F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df1F <- df1F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df1F <- df1F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df2F <- df2 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df2F <- df2F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df2F <- df2F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df2F <- df2F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df2F <- df2F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df3F <- df3 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df3F <- df3F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df3F <- df3F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df3F <- df3F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df3F <- df3F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df4F <- df4 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df4F <- df4F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df4F <- df4F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df4F <- df4F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df4F <- df4F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df5F <- df5 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df5F <- df5F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df5F <- df5F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df5F <- df5F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df5F <- df5F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df6F <- df6 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df6F <- df6F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df6F <- df6F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df6F <- df6F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df6F <- df6F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df7F <- df7 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df7F <- df7F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df7F <- df7F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df7F <- df7F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df7F <- df7F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df8F <- df8 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df8F <- df8F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df8F <- df8F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df8F <- df8F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df8F <- df8F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df9F <- df9 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df9F <- df9F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df9F <- df9F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df9F <- df9F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df9F <- df9F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df10F <- df10 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df10F <- df10F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df10F <- df10F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df10F <- df10F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df10F <- df10F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df11F <- df11 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df11F <- df11F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df11F <- df11F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df11F <- df11F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df11F <- df11F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df12F <- df12 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df12F <- df12F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df12F <- df12F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df12F <- df12F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df12F <- df12F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df13F <- df13 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df13F <- df13F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df13F <- df13F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df13F <- df13F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df13F <- df13F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df14F <- df14 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df14F <- df14F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df14F <- df14F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df14F <- df14F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df14F <- df14F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df15F <- df15 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df15F <- df15F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df15F <- df15F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df15F <- df15F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df15F <- df15F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df16F <- df16 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df16F <- df16F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df16F <- df16F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df16F <- df16F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df16F <- df16F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df17F <- df17 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df17F <- df17F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df17F <- df17F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df17F <- df17F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df17F <- df17F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df18F <- df18 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df18F <- df18F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df18F <- df18F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df18F <- df18F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df18F <- df18F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df19F <- df19 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df19F <- df19F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df19F <- df19F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df19F <- df19F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df19F <- df19F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df20F <- df20 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df20F <- df20F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df20F <- df20F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df20F <- df20F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df20F <- df20F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df21F <- df21 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df21F <- df21F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df21F <- df21F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df21F <- df21F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df21F <- df21F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df22F <- df22 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df22F <- df22F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df22F <- df22F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df22F <- df22F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df22F <- df22F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df23F <- df23 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df23F <- df23F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df23F <- df23F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df23F <- df23F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df23F <- df23F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df24F <- df24 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df24F <- df24F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df24F <- df24F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df24F <- df24F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df24F <- df24F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df25F <- df25 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df25F <- df25F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df25F <- df25F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df25F <- df25F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df25F <- df25F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df26F <- df26 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df26F <- df26F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df26F <- df26F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df26F <- df26F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df26F <- df26F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df27F <- df27 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df27F <- df27F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df27F <- df27F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df27F <- df27F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df27F <- df27F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df28F <- df28 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df28F <- df28F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df28F <- df28F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df28F <- df28F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df28F <- df28F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df29F <- df29 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df29F <- df29F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df29F <- df29F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df29F <- df29F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df29F <- df29F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df30F <- df30 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df30F <- df30F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df30F <- df30F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df30F <- df30F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df30F <- df30F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) df31F <- df31 %>% mutate(MV = factor(MV, levels = c("off", "on"))) df31F <- df31F %>% mutate(Gender = factor(Gender, levels = c("male", "female"))) df31F <- df31F %>% mutate(Mortality = factor(Mortality, levels = c("no", "yes"))) df31F <- df31F %>% mutate(ROX = factor(ROX, levels = c("hi", "lo"))) df31F <- df31F %>% mutate(LIV = factor(LIV, levels = c("hi", "lo"))) |
|
1 2 3 4 5 6 7 |
#install.packages("DataExplorer") #library("DataExplorer") #install.packages("bnlearn") library(bnlearn) #install.packages("BiocManager") #BiocManager::install("Rgraphviz") library(Rgraphviz) |
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 |
dag1 = empty.graph(colnames(df1F)) dag2 = empty.graph(colnames(df2F)) dag3 = empty.graph(colnames(df3F)) dag4 = empty.graph(colnames(df4F)) dag5 = empty.graph(colnames(df5F)) dag6 = empty.graph(colnames(df6F)) dag7 = empty.graph(colnames(df7F)) dag8 = empty.graph(colnames(df8F)) dag9 = empty.graph(colnames(df9F)) dag10 = empty.graph(colnames(df10F)) dag11 = empty.graph(colnames(df11F)) dag12 = empty.graph(colnames(df12F)) dag13 = empty.graph(colnames(df13F)) dag14 = empty.graph(colnames(df14F)) dag15 = empty.graph(colnames(df15F)) dag16 = empty.graph(colnames(df16F)) dag17 = empty.graph(colnames(df17F)) dag18 = empty.graph(colnames(df18F)) dag19 = empty.graph(colnames(df19F)) dag20 = empty.graph(colnames(df20F)) dag21 = empty.graph(colnames(df21F)) dag22 = empty.graph(colnames(df22F)) dag23 = empty.graph(colnames(df23F)) dag24 = empty.graph(colnames(df24F)) dag25 = empty.graph(colnames(df25F)) dag26 = empty.graph(colnames(df26F)) dag27 = empty.graph(colnames(df27F)) dag28 = empty.graph(colnames(df28F)) dag29 = empty.graph(colnames(df29F)) dag30 = empty.graph(colnames(df30F)) dag31 = empty.graph(colnames(df31F)) |
|
1 2 3 4 5 6 |
bl2 = tiers2blacklist(list("Gender", "Age")) bl2 = matrix(c("Gender", "Age", "Mortality","PSI", "Mortality", "Cr"), ncol = 2, byrow = TRUE, dimnames = list(NULL, c("from", "to"))) bl2 |
A matrix: 3 × 2 of type chr
from to
Gender Age
Mortality PSI
Mortality Cr
|
1 2 3 4 5 6 7 8 9 10 11 |
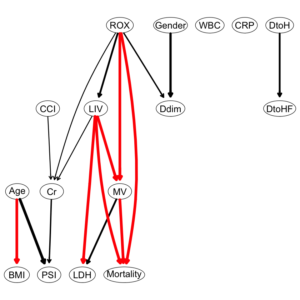
wl2 = matrix(c("ROX", "MV", "LIV", "MV", "LIV", "LDH", "Age", "BMI", "MV", "Mortality", "ROX", "Mortality", "LIV", "Mortality", "Cr", "PSI"), ncol = 2, byrow = TRUE, dimnames = list(NULL, c("from", "to"))) wl2 |
A matrix: 7 × 2 of type chr
from to
ROX MV
LIV MV
LIV LDH
Age BMI
MV Mortality
ROX Mortality
LIV Mortality
ブートストラップ法
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 |
str.raw1_1 = boot.strength(df1F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_1, "threshold") str.raw1_2 = boot.strength(df2F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_2, "threshold") str.raw1_3 = boot.strength(df3F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_3, "threshold") str.raw1_4 = boot.strength(df4F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_4, "threshold") str.raw1_5 = boot.strength(df5F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_5, "threshold") str.raw1_6 = boot.strength(df6F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_6, "threshold") str.raw1_7 = boot.strength(df7F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_7, "threshold") str.raw1_8 = boot.strength(df8F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_8, "threshold") str.raw1_9 = boot.strength(df9F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_9, "threshold") str.raw1_10 = boot.strength(df10F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_10, "threshold") str.raw1_11 = boot.strength(df11F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_11, "threshold") str.raw1_12 = boot.strength(df12F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_12, "threshold") str.raw1_13 = boot.strength(df13F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_13, "threshold") str.raw1_14 = boot.strength(df14F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_14, "threshold") str.raw1_15 = boot.strength(df15F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_15, "threshold") str.raw1_16 = boot.strength(df16F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_16, "threshold") str.raw1_17 = boot.strength(df17F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_17, "threshold") str.raw1_18 = boot.strength(df18F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_18, "threshold") str.raw1_19 = boot.strength(df19F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_19, "threshold") str.raw1_20 = boot.strength(df20F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_20, "threshold") str.raw1_21 = boot.strength(df21F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_21, "threshold") str.raw1_22 = boot.strength(df22F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_22, "threshold") str.raw1_23 = boot.strength(df23F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_23, "threshold") str.raw1_24 = boot.strength(df24F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_24, "threshold") str.raw1_25 = boot.strength(df25F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_25, "threshold") str.raw1_26 = boot.strength(df26F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_26, "threshold") str.raw1_27 = boot.strength(df27F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_27, "threshold") str.raw1_28 = boot.strength(df28F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_28, "threshold") str.raw1_29 = boot.strength(df29F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_29, "threshold") str.raw1_30 = boot.strength(df30F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_30, "threshold") str.raw1_31 = boot.strength(df31F, R = 1000, algorithm = "hc", algorithm.args = list(whitelist = wl2, blacklist = bl2)) attr(str.raw1_31, "threshold") |
0.505
0.505
0.495
0.49
0.5
0.5
0.515
0.5
0.5
0.505
0.5
0.51
0.495
0.495
0.505
0.49
0.5
0.47
0.49
0.5
0.495
0.49
0.505
0.49
0.47
0.55
0.505
0.495
0.49
0.48
0.49
|
1 |
str.raw1_1 |
A bn.strength: 240 × 4
from to strength direction
1 MV ROX 1.000 0.0000000
2 MV LIV 1.000 0.0000000
3 MV Gender 0.040 1.0000000
4 MV Age 0.085 1.0000000
5 MV BMI 0.045 1.0000000
6 MV WBC 0.055 1.0000000
……..
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 |
write.table (str.raw1_1, file = "str.raw1_1.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_2, file = "str.raw1_2.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_3, file = "str.raw1_3.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_4, file = "str.raw1_4.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_5, file = "str.raw1_5.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_6, file = "str.raw1_6.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_7, file = "str.raw1_7.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_8, file = "str.raw1_8.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_9, file = "str.raw1_9.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_10, file = "str.raw1_10.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_11, file = "str.raw1_11.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_12, file = "str.raw1_12.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_13, file = "str.raw1_13.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_14, file = "str.raw1_14.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_15, file = "str.raw1_15.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_16, file = "str.raw1_16.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_17, file = "str.raw1_17.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_18, file = "str.raw1_18.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_19, file = "str.raw1_19.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_20, file = "str.raw1_20.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_21, file = "str.raw1_21.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_22, file = "str.raw1_22.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_23, file = "str.raw1_23.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_24, file = "str.raw1_24.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_25, file = "str.raw1_25.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_26, file = "str.raw1_26.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_27, file = "str.raw1_27.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_28, file = "str.raw1_28.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_29, file = "str.raw1_29.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_30, file = "str.raw1_30.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (str.raw1_31, file = "str.raw1_31.txt", sep = "\t", quote = FALSE, row.names = TRUE) </pre <pre> avg.df1_1 = averaged.network(str.raw1_1) strength.plot(avg.df1_1, str.raw1_1, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_2 = averaged.network(str.raw1_2) strength.plot(avg.df1_2, str.raw1_2, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_3 = averaged.network(str.raw1_3) strength.plot(avg.df1_3, str.raw1_3, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_4 = averaged.network(str.raw1_4) strength.plot(avg.df1_4, str.raw1_4, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_5 = averaged.network(str.raw1_5) strength.plot(avg.df1_5, str.raw1_5, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_6 = averaged.network(str.raw1_6) strength.plot(avg.df1_6, str.raw1_6, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_7 = averaged.network(str.raw1_7) strength.plot(avg.df1_7, str.raw1_7, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_8 = averaged.network(str.raw1_8) strength.plot(avg.df1_8, str.raw1_8, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_9 = averaged.network(str.raw1_9) strength.plot(avg.df1_9, str.raw1_9, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_10 = averaged.network(str.raw1_10) strength.plot(avg.df1_10, str.raw1_10, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_11 = averaged.network(str.raw1_11) strength.plot(avg.df1_11, str.raw1_11, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_12 = averaged.network(str.raw1_12) strength.plot(avg.df1_12, str.raw1_12, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_13 = averaged.network(str.raw1_13) strength.plot(avg.df1_13, str.raw1_13, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_14 = averaged.network(str.raw1_14) strength.plot(avg.df1_14, str.raw1_14, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_15 = averaged.network(str.raw1_15) strength.plot(avg.df1_15, str.raw1_15, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_16 = averaged.network(str.raw1_16) strength.plot(avg.df1_16, str.raw1_16, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_17 = averaged.network(str.raw1_17) strength.plot(avg.df1_17, str.raw1_17, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_18 = averaged.network(str.raw1_18) strength.plot(avg.df1_18, str.raw1_18, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_19 = averaged.network(str.raw1_19) strength.plot(avg.df1_19, str.raw1_19, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_20 = averaged.network(str.raw1_20) strength.plot(avg.df1_20, str.raw1_20, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_21 = averaged.network(str.raw1_21) strength.plot(avg.df1_21, str.raw1_21, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_22 = averaged.network(str.raw1_22) strength.plot(avg.df1_22, str.raw1_22, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_23 = averaged.network(str.raw1_23) strength.plot(avg.df1_23, str.raw1_23, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_24 = averaged.network(str.raw1_24) strength.plot(avg.df1_24, str.raw1_24, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_25 = averaged.network(str.raw1_25) strength.plot(avg.df1_25, str.raw1_25, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_26 = averaged.network(str.raw1_26) strength.plot(avg.df1_26, str.raw1_26, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_27 = averaged.network(str.raw1_27) strength.plot(avg.df1_27, str.raw1_27, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_28 = averaged.network(str.raw1_28) strength.plot(avg.df1_28, str.raw1_28, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_29 = averaged.network(str.raw1_29) strength.plot(avg.df1_29, str.raw1_29, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_30 = averaged.network(str.raw1_30) strength.plot(avg.df1_30, str.raw1_30, shape = "ellipse", highlight = list(arcs = wl2)) avg.df1_31 = averaged.network(str.raw1_31) strength.plot(avg.df1_31, str.raw1_31, shape = "ellipse", highlight = list(arcs = wl2)) |
|
1 |
avg.df1_1 |
Random/Generated Bayesian network
model:
[ROX][Gender][WBC][CRP][DtoH][LIV|ROX][Ddim|Gender][DtoHF|DtoH][MV|ROX:LIV]
[Cr|MV:Gender][LDH|MV:LIV:Ddim][Mortality|MV:ROX:LIV][PSI|LIV:Cr][Age|PSI]
[CCI|PSI][BMI|Age]
nodes: 16
arcs: 18
undirected arcs: 0
directed arcs: 18
average markov blanket size: 2.75
average neighbourhood size: 2.25
average branching factor: 1.12
generation algorithm: Model Averaging
significance threshold: 0.5
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 |
avg.raw.full1_1 = averaged.network(str.raw1_1) avg.raw.full1_2 = averaged.network(str.raw1_2) avg.raw.full1_3 = averaged.network(str.raw1_3) avg.raw.full1_4 = averaged.network(str.raw1_4) avg.raw.full1_5 = averaged.network(str.raw1_5) avg.raw.full1_6 = averaged.network(str.raw1_6) avg.raw.full1_7 = averaged.network(str.raw1_7) avg.raw.full1_8 = averaged.network(str.raw1_8) avg.raw.full1_9 = averaged.network(str.raw1_9) avg.raw.full1_10 = averaged.network(str.raw1_10) avg.raw.full1_11 = averaged.network(str.raw1_11) avg.raw.full1_12 = averaged.network(str.raw1_12) avg.raw.full1_13 = averaged.network(str.raw1_13) avg.raw.full1_14 = averaged.network(str.raw1_14) avg.raw.full1_15 = averaged.network(str.raw1_15) avg.raw.full1_16 = averaged.network(str.raw1_16) avg.raw.full1_17 = averaged.network(str.raw1_17) avg.raw.full1_18 = averaged.network(str.raw1_18) avg.raw.full1_19 = averaged.network(str.raw1_19) avg.raw.full1_20 = averaged.network(str.raw1_20) avg.raw.full1_21 = averaged.network(str.raw1_21) avg.raw.full1_22 = averaged.network(str.raw1_22) avg.raw.full1_23 = averaged.network(str.raw1_23) avg.raw.full1_24 = averaged.network(str.raw1_24) avg.raw.full1_25 = averaged.network(str.raw1_25) avg.raw.full1_26 = averaged.network(str.raw1_26) avg.raw.full1_27 = averaged.network(str.raw1_27) avg.raw.full1_28 = averaged.network(str.raw1_28) avg.raw.full1_29 = averaged.network(str.raw1_29) avg.raw.full1_30 = averaged.network(str.raw1_30) avg.raw.full1_31 = averaged.network(str.raw1_31) |
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 |
strength.plot(avg.raw.full1_1, str.raw1_1, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_2, str.raw1_2, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_3, str.raw1_3, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_4, str.raw1_4, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_5, str.raw1_5, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_6, str.raw1_6, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_7, str.raw1_7, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_8, str.raw1_8, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_9, str.raw1_9, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_10, str.raw1_10, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_11, str.raw1_11, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_12, str.raw1_12, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_13, str.raw1_13, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_14, str.raw1_14, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_15, str.raw1_15, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_16, str.raw1_16, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_17, str.raw1_17, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_18, str.raw1_18, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_19, str.raw1_19, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_20, str.raw1_20, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_21, str.raw1_21, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_22, str.raw1_22, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_23, str.raw1_23, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_24, str.raw1_24, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_25, str.raw1_25, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_26, str.raw1_26, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_27, str.raw1_27, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_28, str.raw1_28, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_29, str.raw1_29, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_30, str.raw1_30, shape = "ellipse", highlight = list(arcs = wl2)) strength.plot(avg.raw.full1_31, str.raw1_31, shape = "ellipse", highlight = list(arcs = wl2)) |
|
1 |
arc.strength(avg.raw.full1_1, df1F) |
A bn.strength: 16 × 3
from to strength
1 MV Cr 1.608868e-03
2 MV LDH 3.341670e-09
3 MV Mortality 3.016962e-01
4 ROX MV 4.080550e-01
5 ROX LIV 3.772321e-03
……
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 |
write.table (arc.strength(avg.raw.full1_1, df1F), file = "avg.raw.full1_1.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_2, df2F), file = "avg.raw.full1_2.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_3, df3F), file = "avg.raw.full1_3.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_4, df4F), file = "avg.raw.full1_4.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_5, df5F), file = "avg.raw.full1_5.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_6, df6F), file = "avg.raw.full1_6.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_7, df7F), file = "avg.raw.full1_7.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_8, df8F), file = "avg.raw.full1_8.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_9, df9F), file = "avg.raw.full1_9.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_10, df10F), file = "avg.raw.full1_10.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_11, df11F), file = "avg.raw.full1_11.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_12, df12F), file = "avg.raw.full1_12.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_13, df13F), file = "avg.raw.full1_13.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_14, df14F), file = "avg.raw.full1_14.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_15, df15F), file = "avg.raw.full1_15.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_16, df16F), file = "avg.raw.full1_16.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_17, df17F), file = "avg.raw.full1_17.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_18, df18F), file = "avg.raw.full1_18.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_19, df19F), file = "avg.raw.full1_19.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_20, df20F), file = "avg.raw.full1_20.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_21, df21F), file = "avg.raw.full1_21.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_22, df22F), file = "avg.raw.full1_22.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_23, df23F), file = "avg.raw.full1_23.txt", sep = "\t", quote = FALSE, row.names = TRUE) #write.table (arc.strength(avg.raw.full1_24, df24F), file = "avg.raw.full1_24.txt", sep = "\t", # quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_25, df25F), file = "avg.raw.full1_25.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_26, df26F), file = "avg.raw.full1_26.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_27, df27F), file = "avg.raw.full1_27.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_28, df28F), file = "avg.raw.full1_28.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_29, df29F), file = "avg.raw.full1_29.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_30, df30F), file = "avg.raw.full1_30.txt", sep = "\t", quote = FALSE, row.names = TRUE) write.table (arc.strength(avg.raw.full1_31, df31F), file = "avg.raw.full1_31.txt", sep = "\t", quote = FALSE, row.names = TRUE) |
推測
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 |
fitted_f16_1 = bn.fit(avg.raw.full1_1, data = df1F) fitted_f16_2 = bn.fit(avg.raw.full1_2, data = df2F) fitted_f16_3 = bn.fit(avg.raw.full1_3, data = df3F) fitted_f16_4 = bn.fit(avg.raw.full1_4, data = df4F) fitted_f16_5 = bn.fit(avg.raw.full1_5, data = df5F) fitted_f16_6 = bn.fit(avg.raw.full1_6, data = df6F) fitted_f16_7 = bn.fit(avg.raw.full1_7, data = df7F) fitted_f16_8 = bn.fit(avg.raw.full1_8, data = df8F) fitted_f16_9 = bn.fit(avg.raw.full1_9, data = df9F) fitted_f16_10 = bn.fit(avg.raw.full1_10, data = df10F) fitted_f16_11 = bn.fit(avg.raw.full1_11, data = df11F) fitted_f16_12 = bn.fit(avg.raw.full1_12, data = df12F) fitted_f16_13 = bn.fit(avg.raw.full1_13, data = df13F) fitted_f16_14 = bn.fit(avg.raw.full1_14, data = df14F) fitted_f16_15 = bn.fit(avg.raw.full1_15, data = df15F) fitted_f16_16 = bn.fit(avg.raw.full1_16, data = df16F) fitted_f16_17 = bn.fit(avg.raw.full1_17, data = df17F) fitted_f16_18 = bn.fit(avg.raw.full1_18, data = df18F) fitted_f16_19 = bn.fit(avg.raw.full1_19, data = df19F) fitted_f16_20 = bn.fit(avg.raw.full1_20, data = df20F) fitted_f16_21 = bn.fit(avg.raw.full1_21, data = df21F) fitted_f16_22 = bn.fit(avg.raw.full1_22, data = df22F) fitted_f16_23 = bn.fit(avg.raw.full1_23, data = df23F) fitted_f16_24 = bn.fit(avg.raw.full1_24, data = df24F) fitted_f16_25 = bn.fit(avg.raw.full1_25, data = df25F) fitted_f16_26 = bn.fit(avg.raw.full1_26, data = df26F) fitted_f16_27 = bn.fit(avg.raw.full1_27, data = df27F) fitted_f16_28 = bn.fit(avg.raw.full1_28, data = df28F) fitted_f16_29 = bn.fit(avg.raw.full1_29, data = df29F) fitted_f16_30 = bn.fit(avg.raw.full1_30, data = df30F) fitted_f16_31 = bn.fit(avg.raw.full1_31, data = df31F) |
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 |
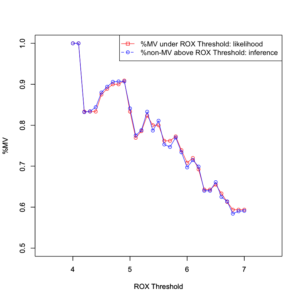
# MV:on, ROX:low mvp1 = round(cpquery(fitted_f16_1, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp2 = round(cpquery(fitted_f16_2, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp3 = round(cpquery(fitted_f16_3, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp4 = round(cpquery(fitted_f16_4, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp5 = round(cpquery(fitted_f16_5, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp6 = round(cpquery(fitted_f16_6, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp7 = round(cpquery(fitted_f16_7, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp8 = round(cpquery(fitted_f16_8, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp9 = round(cpquery(fitted_f16_9, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp10 = round(cpquery(fitted_f16_10, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp11 = round(cpquery(fitted_f16_11, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp12 = round(cpquery(fitted_f16_12, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp13 = round(cpquery(fitted_f16_13, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp14 = round(cpquery(fitted_f16_14, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp15 = round(cpquery(fitted_f16_15, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp16 = round(cpquery(fitted_f16_16, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp17 = round(cpquery(fitted_f16_17, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp18 = round(cpquery(fitted_f16_18, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp19 = round(cpquery(fitted_f16_19, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp20 = round(cpquery(fitted_f16_20, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp21 = round(cpquery(fitted_f16_21, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp22 = round(cpquery(fitted_f16_22, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp23 = round(cpquery(fitted_f16_23, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp24 = round(cpquery(fitted_f16_24, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp25 = round(cpquery(fitted_f16_25, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp26 = round(cpquery(fitted_f16_26, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp27 = round(cpquery(fitted_f16_27, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp28 = round(cpquery(fitted_f16_28, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp29 = round(cpquery(fitted_f16_29, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp30 = round(cpquery(fitted_f16_30, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) mvp31 = round(cpquery(fitted_f16_31, event = (MV == "on"), evidence = (ROX == "lo")), digits = 3) |
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 |
# MV:off, ROX:hi mvq1 = round(cpquery(fitted_f16_1, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq2 = round(cpquery(fitted_f16_2, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq3 = round(cpquery(fitted_f16_3, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq4 = round(cpquery(fitted_f16_4, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq5 = round(cpquery(fitted_f16_5, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq6 = round(cpquery(fitted_f16_6, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq7 = round(cpquery(fitted_f16_7, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq8 = round(cpquery(fitted_f16_8, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq9 = round(cpquery(fitted_f16_9, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq10 = round(cpquery(fitted_f16_10, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq11 = round(cpquery(fitted_f16_11, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq12 = round(cpquery(fitted_f16_12, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq13 = round(cpquery(fitted_f16_13, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq14 = round(cpquery(fitted_f16_14, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq15 = round(cpquery(fitted_f16_15, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq16 = round(cpquery(fitted_f16_16, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq17 = round(cpquery(fitted_f16_17, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq18 = round(cpquery(fitted_f16_18, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq19 = round(cpquery(fitted_f16_19, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq20 = round(cpquery(fitted_f16_20, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq21 = round(cpquery(fitted_f16_21, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq22 = round(cpquery(fitted_f16_22, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq23 = round(cpquery(fitted_f16_23, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq24 = round(cpquery(fitted_f16_24, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq25 = round(cpquery(fitted_f16_25, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq26 = round(cpquery(fitted_f16_26, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq27 = round(cpquery(fitted_f16_27, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq28 = round(cpquery(fitted_f16_28, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq29 = round(cpquery(fitted_f16_29, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq30 = round(cpquery(fitted_f16_30, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) mvq31 = round(cpquery(fitted_f16_31, event = (MV == "off"), evidence = (ROX == "hi")), digits = 3) |
|
1 2 3 4 5 6 7 8 9 |
mvp_all <- c(mvp1, mvp2, mvp3, mvp4, mvp5, mvp6, mvp7, mvp8, mvp9, mvp10, mvp11, mvp12, mvp13, mvp14, mvp15, mvp16, mvp17, mvp18, mvp19, mvp20, mvp21, mvp22, mvp23, mvp24, mvp25, mvp26, mvp27, mvp28, mvp29, mvp30, mvp31) mvq_all <- c(mvq1, mvq2, mvq3, mvq4, mvq5, mvq6, mvq7, mvq8, mvq9, mvq10, mvq11, mvq12, mvq13, mvq14, mvq15, mvq16, mvq17, mvq18, mvq19, mvq20, mvq21, mvq22, mvq23, mvq24, mvq25, mvq26, mvq27, mvq28, mvq29, mvq30, mvq31) |
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 |
mt1 = round(cpquery(fitted_f16_1, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt2 = round(cpquery(fitted_f16_2, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt3= round(cpquery(fitted_f16_3, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt4 = round(cpquery(fitted_f16_4, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt5 = round(cpquery(fitted_f16_5, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt6 = round(cpquery(fitted_f16_6, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt7 = round(cpquery(fitted_f16_7, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt8 = round(cpquery(fitted_f16_8, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt9 = round(cpquery(fitted_f16_9, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt10 = round(cpquery(fitted_f16_10, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt11 = round(cpquery(fitted_f16_11, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt12 = round(cpquery(fitted_f16_12, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt13 = round(cpquery(fitted_f16_13, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt14 = round(cpquery(fitted_f16_14, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt15 = round(cpquery(fitted_f16_15, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt16 = round(cpquery(fitted_f16_16, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt17 = round(cpquery(fitted_f16_17, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt18 = round(cpquery(fitted_f16_18, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt19 = round(cpquery(fitted_f16_19, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt20 = round(cpquery(fitted_f16_20, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt21 = round(cpquery(fitted_f16_21, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt22 = round(cpquery(fitted_f16_22, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt23 = round(cpquery(fitted_f16_23, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt24 = round(cpquery(fitted_f16_24, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt25 = round(cpquery(fitted_f16_25, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt26 = round(cpquery(fitted_f16_26, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt27 = round(cpquery(fitted_f16_27, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt28 = round(cpquery(fitted_f16_28, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt29 = round(cpquery(fitted_f16_29, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt30 = round(cpquery(fitted_f16_30, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) mt31 = round(cpquery(fitted_f16_31, event = (Mortality == "yes"), evidence = (ROX == "lo")), digits = 3) |
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 |
mu1 = round(cpquery(fitted_f16_1, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu2 = round(cpquery(fitted_f16_2, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu3= round(cpquery(fitted_f16_3, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu4 = round(cpquery(fitted_f16_4, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu5 = round(cpquery(fitted_f16_5, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu6 = round(cpquery(fitted_f16_6, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu7 = round(cpquery(fitted_f16_7, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu8 = round(cpquery(fitted_f16_8, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu9 = round(cpquery(fitted_f16_9, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu10 = round(cpquery(fitted_f16_10, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu11 = round(cpquery(fitted_f16_11, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu12 = round(cpquery(fitted_f16_12, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu13 = round(cpquery(fitted_f16_13, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu14 = round(cpquery(fitted_f16_14, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu15 = round(cpquery(fitted_f16_15, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu16 = round(cpquery(fitted_f16_16, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu17 = round(cpquery(fitted_f16_17, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu18 = round(cpquery(fitted_f16_18, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu19 = round(cpquery(fitted_f16_19, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu20 = round(cpquery(fitted_f16_20, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu21 = round(cpquery(fitted_f16_21, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu22 = round(cpquery(fitted_f16_22, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu23 = round(cpquery(fitted_f16_23, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu24 = round(cpquery(fitted_f16_24, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu25 = round(cpquery(fitted_f16_25, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu26 = round(cpquery(fitted_f16_26, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu27 = round(cpquery(fitted_f16_27, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu28 = round(cpquery(fitted_f16_28, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu29 = round(cpquery(fitted_f16_29, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu30 = round(cpquery(fitted_f16_30, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) mu31 = round(cpquery(fitted_f16_31, event = (Mortality == "no"), evidence = (ROX == "hi")), digits = 3) |
|
1 2 3 4 5 6 7 8 9 |
mt_all <- c(mt1, mt2, mt3, mt4, mt5, mt6, mt7, mt8, mt9, mt10, mt11, mt12, mt13, mt14, mt15, mt16, mt17, mt18, mt19, mt20, mt21, mt22, mt23, mt24, mt25, mt26, mt27, mt28, mt29, mt30, mt31) mu_all <- c(mu1, mu2, mu3, mu4, mu5, mu6, mu7, mu8, mu9, mu10, mu11, mu12, mu13, mu14, mu15, mu16, mu17, mu18, mu19, mu20, mu21, mu22, mu23, mu24, mu25, mu26, mu27, mu28, mu29, mu30, mu31) |
|
1 2 |
data3 <- cbind(data, mvp_all, mt_all, mvq_all, mu_all) data3 |
A matrix: 31 × 17
ROX m_all n_all r_all s_all t_all u_all mR_all nR_all rR_all sR_all tR_all uR_all mvp_all mt_all mvq_all mu_all
4 4 4 1 1 4 0.25 35 55 0.6363636 52 55 0.9454545 1 0.209 0.637 0.947
4.1 4 4 1 1 4 0.25 35 55 0.6363636 52 55 0.9454545 1 0.22 0.635 0.948
4.2 5 6 0.8333333 1 6 0.1666667 34 53 0.6415094 50 53 0.9433962 0.832 0.18 0.637 0.944
4.3 5 6 0.8333333 1 6 0.1666667 34 53 0.6415094 50 53 0.9433962 0.834 0.144 0.639 0.946
……..
|
1 2 |
names(data3) <- names(data) <- c("ROX", "m", "n", "r", "s", "t", "u", "mR", "nR", "rR", "sR", "tR", "uR", "mvp", "mt", "mvq", "mu") data3 |
|
1 2 3 4 |
plot(data3[,1], data3[,4], type = "o", xlim =c(3.5,7.5), ylim=c(0.5, 1), col="red", xlab="ROX Threshold", ylab="%MV") par(new=T) plot(data3[,1], data3[,14], type = "o", xlim =c(3.5,7.5), ylim=c(0.5, 1), col="blue", xlab="ROX Threshold", ylab="%MV") legend("topright", legend = c("%MV under ROX Threshold: likelihood","%non-MV above ROX Threshold: inference"), col = c("red", "blue"), pch = c(0, 1),lty = c("solid", "dashed")) |
|
1 2 3 4 |
plot(data3[,1], data3[,7], type = "o", xlim =c(3.5,7.5), ylim=c(0, 0.3), col="red", xlab="ROX Threshold", ylab="%Mortality") par(new=T) plot(data3[,1], data3[,15], type = "o", xlim =c(3.5,7.5), ylim=c(0, 0.3), col="blue", xlab="ROX Threshold", ylab="%Mortality") legend("topright", legend = c("%Mortality under ROX Threshold: likelihood","%Mortality above ROX Threshold: inference"), col = c("red", "blue"), pch = c(0, 1),lty = c("solid", "dashed")) |
|
1 2 3 4 |
plot(data3[,1], data3[,14], type = "o", xlim =c(3.5,7.5), ylim=c(0, 1.0), col="red", xlab="ROX Threshold", ylab="case(%)") par(new=T) plot(data3[,1], data3[,16], type = "o", xlim =c(3.5,7.5), ylim=c(0, 1.0), col="blue", xlab="ROX Threshold", ylab="case(%)") legend("bottomright", legend = c("%MV under ROX Threshold: inference","%non-MV above ROX Threshold: inference"), col = c("red", "blue"), pch = c(0, 1),lty = c("solid", "dashed")) |
|
1 2 3 4 |
plot(data3[,1], data3[,15], type = "o", xlim =c(3.5,7.5), ylim=c(0, 1.0), col="red", xlab="ROX Threshold", ylab="case(%)") par(new=T) plot(data3[,1], data3[,17], type = "o", xlim =c(3.5,7.5), ylim=c(0, 1.0), col="blue", xlab="ROX Threshold", ylab="case(%)") legend("center", legend = c("%Mortality under ROX Threshold: inderence","%Surival above ROX Threshold: inference"), col = c("red", "blue"), pch = c(0, 1),lty = c("solid", "dashed")) |