????????????
ベイズ推論による機械学習入門 須山 敦志 著 を読む
????????????
Bayesian Poisson HMM
????????????
いよいよ最後、Bayesian Poisson HMMは、貼付の自作モジュール:PoissonHMM.jlとPoissonMixtureModel.jlと2つをimportする。
加えて、HDF5でエラーデまくりで、とにかく、Mac OS X環境にHDF5インストール。
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 |
MacBook-Pro-6:~ $ brew install hdf5 Updating Homebrew... ==> Auto-updated Homebrew! Updated 2 taps (homebrew/core and homebrew/cask). ==> Updated Formulae gauge ldc tomee-webprofile jvgrep openapi-generator xonsh Error: Could not link: /usr/local/share/man/man1/brew.1 Please delete these paths and run `brew update`. ==> Installing dependencies for hdf5: szip ==> Installing hdf5 dependency: szip ==> Downloading https://homebrew.bintray.com/bottles/szip-2.1.1_1.mojave.bottle. ######################################################################## 100.0% ==> Pouring szip-2.1.1_1.mojave.bottle.tar.gz 🍺 /usr/local/Cellar/szip/2.1.1_1: 11 files, 109.4KB ==> Installing hdf5 ==> Downloading https://homebrew.bintray.com/bottles/hdf5-1.10.5_1.mojave.bottle ==> Downloading from https://akamai.bintray.com/28/28ee1944f9b17a50bddbfbc1730d0 ######################################################################## 100.0% ==> Pouring hdf5-1.10.5_1.mojave.bottle.tar.gz Warning: hdf5 dependency gcc was built with a different C++ standard library (libstdc++ from clang). This may cause problems at runtime. 🍺 /usr/local/Cellar/hdf5/1.10.5_1: 256 files, 14.6MB MacBook-Pro-6:~ $ |
関係があったのか、なかったのかようわからんが、以下のように、
まずは、Bayesian 1dim Poisson Hidden Markov Model PoissonHMM.jlを入力
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 |
""" Bayesian 1dim Poisson Hidden Markov Model """ module PoissonHMM using StatsFuns.logsumexp, SpecialFunctions.digamma using Distributions export Gam, GHMM, Poi, HMM export sample_HMM, sample_data, winner_takes_all export learn_VI #################### ## Types struct Gam # Parameters of Gamma distribution # 1dim a::Float64 b::Float64 end 途中省略..... function learn_VI(X::Vector{Float64}, prior_bhmm::BHMM, max_iter::Int) # initialisation expt_Z, expt_ZZ = init_Z(X, prior_bhmm) bhmm = add_stats(prior_bhmm, X, expt_Z, expt_ZZ) VB = NaN * zeros(max_iter) # inference for i in 1 : max_iter # E-step #expt_Z, expt_ZZ = update_Z(bhmm, X, expt_Z) expt_Z, expt_ZZ = update_Z_fb(bhmm, X) # M-step bhmm = add_stats(prior_bhmm, X, expt_Z, expt_ZZ) end return expt_Z, bhmm end end |
Out: PoissonHMM
|
1 |
using PoissonHMM |
次に、PoissonMixtureModel.jlを入力
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 |
""" Bayesian Poisson Mixture Model """ module PoissonMixtureModel using StatsFuns.logsumexp, SpecialFunctions.digamma using Distributions export Gam, BPMM, Poi, PMM export sample_PMM, sample_data, winner_takes_all export learn_GS, learn_CGS, learn_VI 途中省略.... """ Compute posterior distribution via collapsed Gibbs sampling. """ function learn_CGS(X::Matrix{Float64}, prior_bpmm::BPMM, max_iter::Int) # initialisation S = init_S(X, prior_bpmm) bpmm = add_stats(prior_bpmm, X, S) VB = NaN * zeros(max_iter) # inference for i in 1 : max_iter # directly sample S S, bpmm = sample_S_CGS(S, X, bpmm) # calc VB VB[i] = calc_ELBO(X, prior_bpmm, bpmm) end return S, bpmm, VB end end |
Out: PoissonMixtureModel
|
1 |
using PoissonMixtureModel |
で、demo_PoissonHMM.jlを入力、実行へ:
|
1 2 3 4 5 |
################################### ## Example code ## for Bayesian Poisson HMM using PyPlot, PyCall |
using HDF5, JLD
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
Out: INFO: Precompiling module HDF5. WARNING: uuid4(rng) is deprecated, use Compat.UUIDs.uuid4(rng) instead. Stacktrace: [1] depwarn(::String, ::Symbol) at ./deprecated.jl:70 [2] uuid4(::MersenneTwister) at ./deprecated.jl:57 [3] msg_header at /Users/teijisw/.julia/v0.6/IJulia/src/msg.jl:18 [inlined] [4] msg_pub(::IJulia.Msg, ::String, ::Dict{String,String}, ::Dict{String,Any}) at /Users/teijisw/.julia/v0.6/IJulia/src/msg.jl:30 (repeats 2 times) [5] send_stream(::String) at /Users/teijisw/.julia/v0.6/IJulia/src/stdio.jl:172 [6] send_stdio(::String) at /Users/teijisw/.julia/v0.6/IJulia/src/stdio.jl:130 [7] (::Base.##302#303{IJulia.#send_stderr,Timer})() at ./event.jl:436 while loading In[6], in expression starting on line 1 INFO: Precompiling module JLD. と一杯警告がでるが、なんとか突破。 <pre> @pyimport matplotlib.gridspec as gspec |
ロードするプログラムの絶対パスに注意して:
|
1 2 3 |
push!(LOAD_PATH,"/Users/******/Desktop/BayesBook-master/src") import PoissonHMM import PoissonMixtureModel |
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 |
""" Simple comparison between HMM and mixture model. """ function test_comparison() ######################### ## load data file_name = "/Users/********/Desktop/BayesBook-master/data/timeseries.jld" X = load(file_name)["obs"] N = length(X) ######################### ## Poison HMM ## set model K = 2 # number of mixture components alpha_phi = 10.0 * ones(K) alpha_A = 100.0 * eye(K) + 1.0*ones(K, K) cmp = [PoissonHMM.Gam(1.0, 0.01), PoissonHMM.Gam(1.0, 0.01)] bhmm = PoissonHMM.BHMM(K, alpha_phi, alpha_A, cmp) ## inference max_iter = 100 tic() Z_est_hmm, post_bhmm = PoissonHMM.learn_VI(X, bhmm, max_iter) toc() ######################### ## Poison Mixture Model ## set model K = 2 # number of mixture components alpha_phi = 10.0 * ones(K) cmp = [PoissonMixtureModel.Gam([1.0], 0.01), PoissonMixtureModel.Gam([1.0], 0.01)] bpmm = PoissonMixtureModel.BPMM(1, K, alpha_phi, cmp) ## inference max_iter = 100 tic() Z_est_pmm, post_bpmm = PoissonMixtureModel.learn_VI(reshape(X, 1, N), bpmm, max_iter) toc() ######################### ## Compare results figure("Hidden Markov Model vs Mixture Model") subplot(3,1,1);plot(X);ylabel("data") subplot(3,1,2);fill_between(1:N, reshape(Z_est_hmm[1,:]', N), zeros(N));ylim([0.0, 1.0]);ylabel("S (PHMM)") subplot(3,1,3);fill_between(1:N, reshape(Z_est_pmm[1,:]', N), zeros(N));ylim([0.0, 1.0]);ylabel("S (PMM)") show() end |
Out: test_comparison
|
1 |
test_comparison() |
elapsed time: 1.493772971 seconds
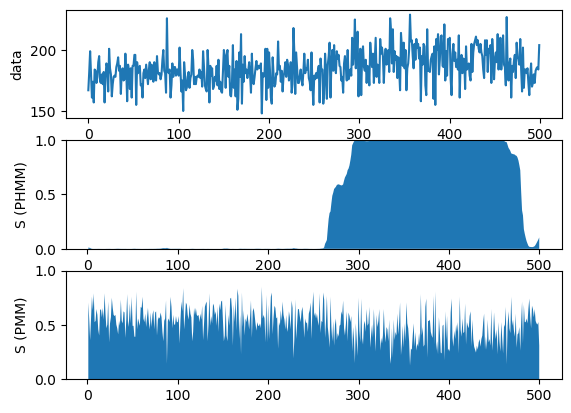
elapsed time: 1.195815636 seconds
ここまでで気がついたが、Julia Ver0.6で Jupyterで学習デモのプログラムを実行させるときは、要するに自作モジュールを使うときも
|
1 2 3 |
push!(LOAD_PATH,"/Users/********/Desktop/BayesBook-master/src") import PoissonHMM import PoissonMixtureModel |
で直接、指定パスからJuliaのモジュールを導入しているので、これまで行ってきたように事前にモジュールを入力実行してusing宣言しなくとも、いきなりデモプログラムを入力実行でよい。
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 |
""" Simple comparison between HMM and mixture model. """ function test_comparison() ######################### ## load data file_name = "/Users/********/Desktop/BayesBook-master/data/timeseries.jld" X = load(file_name)["obs"] N = length(X) ######################### ## Poison HMM ## set model K = 2 # number of mixture components alpha_phi = 10.0 * ones(K) alpha_A = 100.0 * eye(K) + 1.0*ones(K, K) cmp = [PoissonHMM.Gam(1.0, 0.01), PoissonHMM.Gam(1.0, 0.01)] bhmm = PoissonHMM.BHMM(K, alpha_phi, alpha_A, cmp) ## inference max_iter = 100 tic() Z_est_hmm, post_bhmm = PoissonHMM.learn_VI(X, bhmm, max_iter) toc() ######################### ## Poison Mixture Model ## set model K = 2 # number of mixture components alpha_phi = 10.0 * ones(K) cmp = [PoissonMixtureModel.Gam([1.0], 0.01), PoissonMixtureModel.Gam([1.0], 0.01)] bpmm = PoissonMixtureModel.BPMM(1, K, alpha_phi, cmp) ## inference max_iter = 100 tic() Z_est_pmm, post_bpmm = PoissonMixtureModel.learn_VI(reshape(X, 1, N), bpmm, max_iter) toc() ######################### ## Compare results figure("Hidden Markov Model vs Mixture Model") subplot(3,1,1);plot(X);ylabel("data") subplot(3,1,2);fill_between(1:N, reshape(Z_est_hmm[1,:]', N), zeros(N));ylim([0.0, 1.0]);ylabel("S (PHMM)") subplot(3,1,3);fill_between(1:N, reshape(Z_est_pmm[1,:]', N), zeros(N));ylim([0.0, 1.0]);ylabel("S (PMM)") show() end |
test_comparison
|
1 |
test_comparison() |
Out:
elapsed time: 1.614081116 seconds
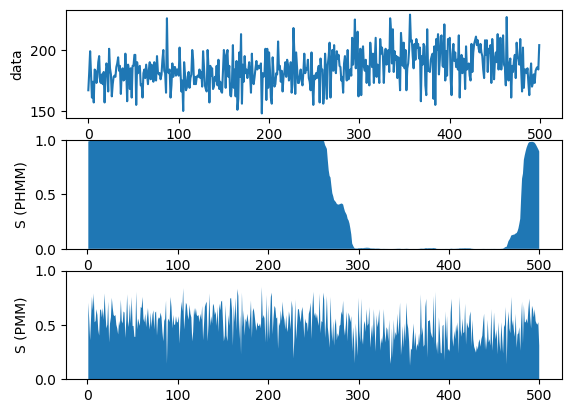
elapsed time: 1.389467078 seconds
で良いわけだ。